| [1] |
MUNDI MS, VELAPATI S, PATEL J, et al. Evolution of NAFLD and its management[J]. Nutr Clin Pract, 2020, 35( 1): 72- 84. DOI: 10.1002/ncp.10449. |
| [2] |
KAUFMANN B, RECA A, WANG B, et al. Mechanisms of nonalcoholic fatty liver disease and implications for surgery[J]. Langenbecks Arch Surg, 2021, 406( 1): 1- 17. DOI: 10.1007/s00423-020-01965-1. |
| [3] |
POUWELS S, SAKRAN N, GRAHAM Y, et al. Non-alcoholic fatty liver disease(NAFLD): A review of pathophysiology, clinical management and effects of weight loss[J]. BMC Endocr Disord, 2022, 22( 1): 63. DOI: 10.1186/s12902-022-00980-1. |
| [4] |
FRIEDMAN SL, NEUSCHWANDER-TETRI BA, RINELLA M, et al. Mechanisms of NAFLD development and therapeutic strategies[J]. Nat Med, 2018, 24( 7): 908- 922. DOI: 10.1038/s41591-018-0104-9. |
| [5] |
XU XH, POULSEN KL, WU LJ, et al. Targeted therapeutics and novel signaling pathways in non-alcohol-associated fatty liver/steatohepatitis(NAFL/NASH)[J]. Signal Transduct Target Ther, 2022, 7( 1): 287. DOI: 10.1038/s41392-022-01119-3. |
| [6] |
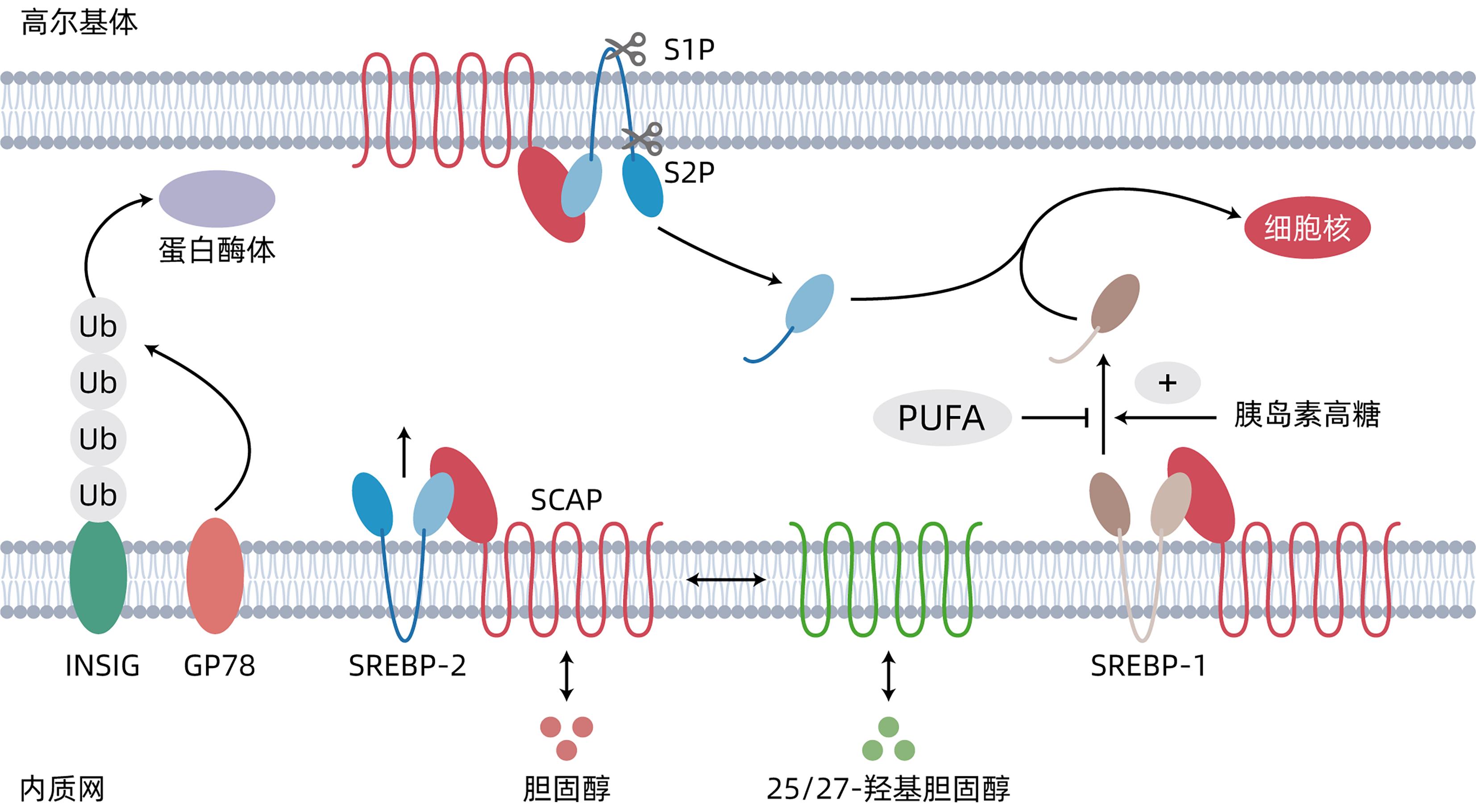
GOLDSTEIN JL, DEBOSE-BOYD RA, BROWN MS. Protein sensors for membrane sterols[J]. Cell, 2006, 124( 1): 35- 46. DOI: 10.1016/j.cell.2005.12.022. |
| [7] |
ARAKI M, NAKAGAWA Y, SAITO H, et al. Hepatocyte- or macrophage-specific SREBP-1a deficiency in mice exacerbates methionine- and choline-deficient diet-induced nonalcoholic fatty liver disease[J]. Am J Physiol Gastrointest Liver Physiol, 2022, 323( 6): G627- G639. DOI: 10.1152/ajpgi.00090.2022. |
| [8] |
SATO R. Sterol metabolism and SREBP activation[J]. Arch Biochem Biophys, 2010, 501( 2): 177- 181. DOI: 10.1016/j.abb.2010.06.004. |
| [9] |
NAKAGAWA Y, ARAKI M, HAN SI, et al. CREBH systemically regulates lipid metabolism by modulating and integrating cellular functions[J]. Nutrients, 2021, 13( 9): 3204. DOI: 10.3390/nu13093204. |
| [10] |
JEON YG, KIM YY, LEE G, et al. Physiological and pathological roles of lipogenesis[J]. Nat Metab, 2023, 5( 5): 735- 759. DOI: 10.1038/s42255-023-00786-y. |
| [11] |
MARGERIE D, LEFEBVRE P, RAVERDY V, et al. Hepatic transcriptomic signatures of statin treatment are associated with impaired glucose homeostasis in severely obese patients[J]. BMC Med Genomics, 2019, 12( 1): 80. DOI: 10.1186/s12920-019-0536-1. |
| [12] |
IPSEN DH, LYKKESFELDT J, TVEDEN-NYBORG P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease[J]. Cell Mol Life Sci, 2018, 75( 18): 3313- 3327. DOI: 10.1007/s00018-018-2860-6. |
| [13] |
GOLDSTEIN JL, BASU SK, BROWN MS. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells[J]. Methods Enzymol, 1983, 98: 241- 260. DOI: 10.1016/0076-6879(83)98152-1. |
| [14] |
NOHTURFFT A, DEBOSE-BOYD RA, SCHEEK S, et al. Sterols regulate cycling of SREBP cleavage-activating protein(SCAP) between endoplasmic reticulum and Golgi[J]. Proc Natl Acad Sci U S A, 1999, 96( 20): 11235- 11240. DOI: 10.1073/pnas.96.20.11235. |
| [15] |
HORTON JD, COHEN JC, HOBBS HH. Molecular biology of PCSK9: Its role in LDL metabolism[J]. Trends Biochem Sci, 2007, 32( 2): 71- 77. DOI: 10.1016/j.tibs.2006.12.008. |
| [16] |
MUSSO G, GAMBINO R, CASSADER M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis[J]. Prog Lipid Res, 2013, 52( 1): 175- 191. DOI: 10.1016/j.plipres.2012.11.002. |
| [17] |
CAROTTI S, AQUILANO K, ZALFA F, et al. Lipophagy impairment is associated with disease progression in NAFLD[J]. Front Physiol, 2020, 11: 850. DOI: 10.3389/fphys.2020.00850. |
| [18] |
ALLAIRE M, RAUTOU PE, CODOGNO P, et al. Autophagy in liver diseases: Time for translation?[J]. J Hepatol, 2019, 70( 5): 985- 998. DOI: 10.1016/j.jhep.2019.01.026. |
| [19] |
NGUYEN TTP, KIM DY, LEE YG, et al. SREBP-1c impairs ULK1 sulfhydration-mediated autophagic flux to promote hepatic steatosis in high-fat-diet-fed mice[J]. Mol Cell, 2021, 81( 18): 3820- 3832. DOI: 10.1016/j.molcel.2021.06.003. |
| [20] |
NGUYEN TTP, KIM DY, IM SS, et al. Impairment of ULK1 sulfhydration-mediated lipophagy by SREBF1/SREBP-1c in hepatic steatosis[J]. Autophagy, 2021, 17( 12): 4489- 4490. DOI: 10.1080/15548627.2021.1968608. |
| [21] |
IM SS, OSBORNE TF. Protection from bacterial-toxin-induced apoptosis in macrophages requires the lipogenic transcription factor sterol regulatory element binding protein 1a[J]. Mol Cell Biol, 2012, 32( 12): 2196- 2202. DOI: 10.1128/MCB.06294-11. |
| [22] |
LU D, PARISI LR, GOKCUMEN O, et al. SREBP activation contributes to fatty acid accumulations in necroptosis[J]. RSC Chem Biol, 2023, 4( 4): 310- 322. DOI: 10.1039/d2cb00172a. |
| [23] |
GUO CS, CHI ZX, JIANG DL, et al. Cholesterol homeostatic regulator SCAP-SREBP2 integrates NLRP3 inflammasome activation and cholesterol biosynthetic signaling in macrophages[J]. Immunity, 2018, 49( 5): 842- 856. DOI: 10.1016/j.immuni.2018.08.021. |
| [24] |
PETTINELLI P, DEL POZO T, ARAYA J, et al. Enhancement in liver SREBP-1c/PPAR-alpha ratio and steatosis in obese patients: Correlations with insulin resistance and n-3 long-chain polyunsaturated fatty acid depletion[J]. Biochim Biophys Acta, 2009, 1792( 11): 1080- 1086. DOI: 10.1016/j.bbadis.2009.08.015. |
| [25] |
FUJII N, NARITA T, OKITA N, et al. Sterol regulatory element-binding protein-1c orchestrates metabolic remodeling of white adipose tissue by caloric restriction[J]. Aging Cell, 2017, 16( 3): 508- 517. DOI: 10.1111/acel.12576. |
| [26] |
SOUNDARARAJAN R, WISHART AD, VASANTHA RUPASINGHE HP, et al. Quercetin 3-glucoside protects neuroblastoma(SH-SY5Y) cells in vitro against oxidative damage by inducing sterol regulatory element-binding protein-2-mediated cholesterol biosynthesis[J]. J Biol Chem, 2008, 283( 4): 2231- 2245. DOI: 10.1074/jbc.M703583200. |
| [27] |
SHIMANO H, SATO R. SREBP-regulated lipid metabolism: Convergent physiology-divergent pathophysiology[J]. Nat Rev Endocrinol, 2017, 13( 12): 710- 730. DOI: 10.1038/nrendo.2017.91. |
| [28] |
DÜVEL K, YECIES JL, MENON S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1[J]. Mol Cell, 2010, 39( 2): 171- 183. DOI: 10.1016/j.molcel.2010.06.022. |
| [29] |
KOHJIMA M, HIGUCHI N, KATO M, et al. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease[J]. Int J Mol Med, 2008, 21( 4): 507- 511.
|
| [30] |
PUIGSERVER P, RHEE J, DONOVAN J, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction[J]. Nature, 2003, 423( 6939): 550- 555. DOI: 10.1038/nature01667. |
| [31] |
JUMP DB. Dietary polyunsaturated fatty acids and regulation of gene transcription[J]. Curr Opin Lipidol, 2002, 13( 2): 155- 164. DOI: 10.1097/00041433-200204000-00007. |
| [32] |
GNONI A, GIUDETTI AM. Dietary long-chain unsaturated fatty acids acutely and differently reduce the activities of lipogenic enzymes and of citrate carrier in rat liver[J]. J Physiol Biochem, 2016, 72( 3): 485- 494. DOI: 10.1007/s13105-016-0495-3. |
| [33] |
DONG QM, MAJUMDAR G, O’MEALLY RN, et al. Insulin-induced de novo lipid synthesis occurs mainly via mTOR-dependent regulation of proteostasis of SREBP-1c[J]. Mol Cell Biochem, 2020, 463( 1-2): 13- 31. DOI: 10.1007/s11010-019-03625-5. |
| [34] |
FLISTER KFT, PINTO BAS, FRANÇA LM, et al. Long-term exposure to high-sucrose diet down-regulates hepatic endoplasmic reticulum-stress adaptive pathways and potentiates de novo lipogenesis in weaned male mice[J]. J Nutr Biochem, 2018, 62: 155- 166. DOI: 10.1016/j.jnutbio.2018.09.007. |
| [35] |
YE J, RAWSON RB, KOMURO R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs[J]. Mol Cell, 2000, 6( 6): 1355- 1364. DOI: 10.1016/s1097-2765(00)00133-7. |
| [36] |
LEE JS, ZHENG Z, MENDEZ R, et al. Pharmacologic ER stress induces non-alcoholic steatohepatitis in an animal model[J]. Toxicol Lett, 2012, 211( 1): 29- 38. DOI: 10.1016/j.toxlet.2012.02.017. |
| [37] |
RÖHRL C, EIGNER K, WINTER K, et al. Endoplasmic reticulum stress impairs cholesterol efflux and synthesis in hepatic cells[J]. J Lipid Res, 2014, 55( 1): 94- 103. DOI: 10.1194/jlr.M043299. |
| [38] |
GILARDI F, MIGLIAVACCA E, NALDI A, et al. Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals[J]. PLoS Genet, 2014, 10( 3): e1004155. DOI: 10.1371/journal.pgen.1004155. |
| [39] |
MARTELOT GL, CLAUDEL T, GATFIELD D, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis[J]. PLoS Biol, 2009, 7( 9): e1000181. DOI: 10.1371/journal.pbio.1000181. |
| [40] |
GUAN DY, BAE H, ZHOU DS, et al. Hepatocyte SREBP signaling mediates clock communication within the liver[J]. J Clin Invest, 2023, 133( 8): e163018. DOI: 10.1172/JCI163018. |
| [41] |
CHEN M, LIN YK, DANG YK, et al. Reprogramming of rhythmic liver metabolism by intestinal clock[J]. J Hepatol, 2023, 79( 3): 741- 757. DOI: 10.1016/j.jhep.2023.04.040. |
| [42] |
SHAO W, MACHAMER CE, ESPENSHADE PJ. Fatostatin blocks ER exit of SCAP but inhibits cell growth in a SCAP-independent manner[J]. J Lipid Res, 2016, 57( 8): 1564- 1573. DOI: 10.1194/jlr.M069583. |
| [43] |
LI YH, LUAN YS, LI JN, et al. Exosomal miR-199a-5p promotes hepatic lipid accumulation by modulating MST1 expression and fatty acid metabolism[J]. Hepatol Int, 2020, 14( 6): 1057- 1074. DOI: 10.1007/s12072-020-10096-0. |
| [44] |
LI YH, NIE JJ, YANG YH, et al. Redox-unlockable nanoparticle-based MST1 delivery system to attenuate hepatic steatosis via the AMPK/SREBP-1c signaling axis[J]. ACS Appl Mater Interfaces, 2022, 14( 30): 34328- 34341. DOI: 10.1021/acsami.2c05889. |
| [45] |
ASANO L, WATANABE M, RYODEN Y, et al. Vitamin D metabolite, 25-hydroxyvitamin D, regulates lipid metabolism by inducing degradation of SREBP/SCAP[J]. Cell Chem Biol, 2017, 24( 2): 207- 217. DOI: 10.1016/j.chembiol.2016.12.017. |
| [46] |
KAWAGOE F, MENDOZA A, HAYATA Y, et al. Discovery of a vitamin D receptor-silent vitamin D derivative that impairs sterol regulatory element-binding protein in vivo[J]. J Med Chem, 2021, 64( 9): 5689- 5709. DOI: 10.1021/acs.jmedchem.0c02179. |
| [47] |
ISHIBASHI H, NAKAGAWA K, ONIMARU M, et al. Sp1 decoy transfected to carcinoma cells suppresses the expression of vascular endothelial growth factor, transforming growth factor beta1, and tissue factor and also cell growth and invasion activities[J]. Cancer Res, 2000, 60( 22): 6531- 6536.
|
| [48] |
AN HJ, KIM JY, GWON MG, et al. Beneficial effects of SREBP decoy oligodeoxynucleotide in an animal model of hyperlipidemia[J]. Int J Mol Sci, 2020, 21( 2): 552. DOI: 10.3390/ijms21020552. |
| [49] |
JIANG SY, YANG XL, YANG ZM, et al. Discovery of an insulin-induced gene binding compound that ameliorates nonalcoholic steatohepatitis by inhibiting sterol regulatory element-binding protein-mediated lipogenesis[J]. Hepatology, 2022, 76( 5): 1466- 1481. DOI: 10.1002/hep.32381. |
| [50] |
WANG YD, ZHANG JN, XU Z, et al. Identification and action mechanism of lipid regulating components from Rhei Radix et rhizoma[J]. J Ethnopharmacol, 2022, 292: 115179. DOI: 10.1016/j.jep.2022.115179. |
| [51] |
SU ZL, HANG PZ, HU J, et al. Aloe-emodin exerts cholesterol-lowering effects by inhibiting proprotein convertase subtilisin/kexin type 9 in hyperlipidemic rats[J]. Acta Pharmacol Sin, 2020, 41( 8): 1085- 1092. DOI: 10.1038/s41401-020-0392-8. |
| [52] |
MIYATA S, KODAKA M, KIKUCHI A, et al. Sulforaphane suppresses the activity of sterol regulatory element-binding proteins(SREBPs) by promoting SREBP precursor degradation[J]. Sci Rep, 2022, 12( 1): 8715. DOI: 10.1038/s41598-022-12347-6. |
| [53] |
CHO WK, LEE MM, MA JY. Antiviral effect of isoquercitrin against influenza A viral infection via modulating hemagglutinin and neuraminidase[J]. Int J Mol Sci, 2022, 23( 21): 13112. DOI: 10.3390/ijms232113112. |
| [54] |
KIM SH, YUN C, KWON D, et al. Effect of isoquercitrin on free fatty acid-induced lipid accumulation in HepG2 cells[J]. Molecules, 2023, 28( 3): 1476. DOI: 10.3390/molecules28031476. |
| [55] |
NAJAFI-SHOUSHTARI SH, KRISTO F, LI YX, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis[J]. Science, 2010, 328( 5985): 1566- 1569. DOI: 10.1126/science.1189123. |
| [56] |
HORIE T, NISHINO T, BABA O, et al. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice[J]. Nat Commun, 2013, 4: 2883. DOI: 10.1038/ncomms3883. |
| [57] |
LI LF, ZHANG XY, REN HJ, et al. MiR-23a/b-3p promotes hepatic lipid accumulation by regulating Srebp-1c and Fas[J]. J Mol Endocrinol, 2021, 68( 1): 35- 49. DOI: 10.1530/JME-20-0324. |
| [58] |
SOBKY SA EL, ABOUD NK, ASSALY NM EL, et al. Regulation of lipid droplet(LD) formation in hepatocytes via regulation of SREBP1c by non-coding RNAs[J]. Front Med, 2022, 9: 903856. DOI: 10.3389/fmed.2022.903856. |
| [59] |
CHENG CW, DENG XL, XU KS. Increased expression of sterol regulatory element binding protein-2 alleviates autophagic dysfunction in NAFLD[J]. Int J Mol Med, 2018, 41( 4): 1877- 1886. DOI: 10.3892/ijmm.2018.3389. |
| [60] |
DENG XL, PAN XL, CHENG CW, et al. Regulation of SREBP-2 intracellular trafficking improves impaired autophagic flux and alleviates endoplasmic reticulum stress in NAFLD[J]. Biochim Biophys Acta Mol Cell Biol Lipids, 2017, 1862( 3): 337- 350. DOI: 10.1016/j.bbalip.2016.12.007. |
| [61] |
JU UI, JEONG DW, SEO J, et al. Neddylation of sterol regulatory element-binding protein 1c is a potential therapeutic target for nonalcoholic fatty liver treatment[J]. Cell Death Dis, 2020, 11( 4): 283. DOI: 10.1038/s41419-020-2472-6. |
| [62] |
SU SY, TIAN HM, JIA X, et al. Mechanistic insights into the effects of SREBP1c on hepatic stellate cell and liver fibrosis[J]. J Cell Mol Med, 2020, 24( 17): 10063- 10074. DOI: 10.1111/jcmm.15614. |
| [63] |
KAWAMURA S, MATSUSHITA Y, KUROSAKI S, et al. Inhibiting SCAP/SREBP exacerbates liver injury and carcinogenesis in murine nonalcoholic steatohepatitis[J]. J Clin Invest, 2022, 132( 11): e151895. DOI: 10.1172/JCI151895. |







 DownLoad:
DownLoad: