NCEH1基因在肝癌组织与细胞系中的表达及生物学作用
DOI: 10.3969/j.issn.1001-5256.2021.08.023
利益冲突声明: 本研究不存在研究者、伦理委员会成员、受试者监护人以及与公开研究成果有关的利益冲突。
作者贡献声明: 曾裕文负责资料分析、拟定写作思路、收集数据、撰写论文、修改论文; 张芳雍、谭国钳参与指导撰写文章、收集数据、修改论文; 吴帆负责课题设计, 指导撰写文章并最后定稿。
Expression and biological role of the neutral cholesterol ester hydrolase 1 gene in liver cancer tissue and cell lines
-
摘要:
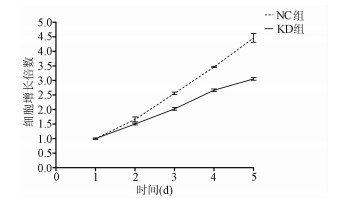
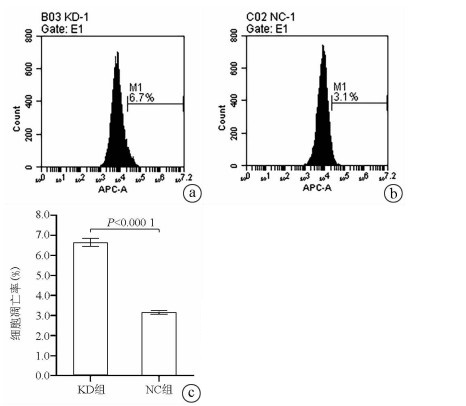
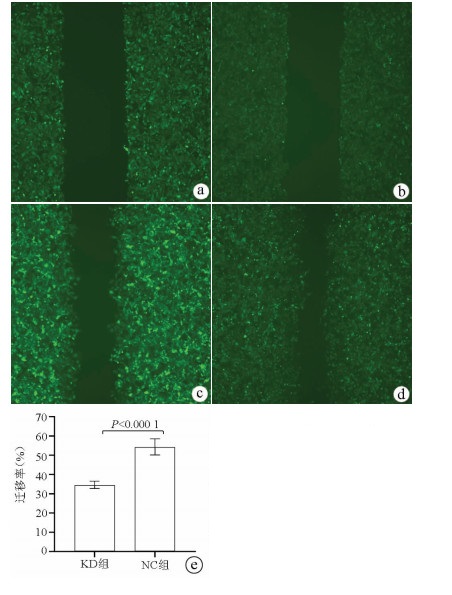
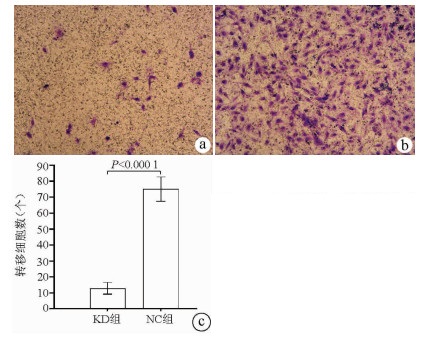
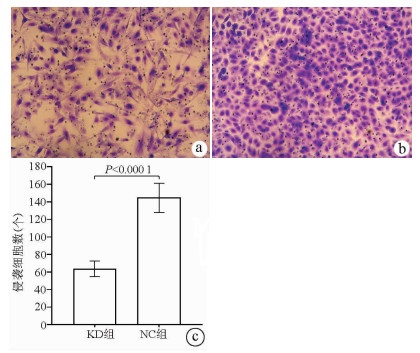
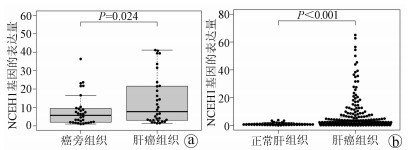
目的 了解中性胆固醇酯水解酶1(neutral cholesterol ester hydrolase 1, NCEH1)基因在肝癌组织及人肝癌细胞系中表达水平, 观察NCEH1基因敲减对SMMC-7721人肝癌细胞系增殖、凋亡、侵袭及转移能力的影响。 方法 选取2013年1月—2019年6月在暨南大学附属广州红十字会医院手术治疗的32例肝癌患者标本及对应的癌旁组织, 采用实时荧光定量PCR方法检测NCEH1基因的相对表达量。从ICGC数据库下载截至2020年9月份的肝癌样本基因表达数据, 应用R软件整理数据, 筛选出每个样本中NCEH1基因表达量, 分别采用配对Wilcoxon符号秩检验和Wilcoxon秩和检验分析肝癌与癌旁组织间的差异。采用实时荧光定量PCR方法检测NCEH1基因在SMMC-7721、Bel-7402、HepG2、Hep3B人肝癌细胞系和HL7702正常人肝细胞系中的表达水平。通过慢病毒介导的小干扰RNA(siRNA)技术构建NCEH1基因敲减的SMMC-7721人肝癌细胞系, 分为NCEH1敲减组(KD组)和阴性对照组(NC组), 以实时荧光定量PCR法检测NCEH1基因的敲减效率, 再以MTT检测实验、Annexin V-APC单染法流式细胞仪检测、划痕愈合实验、Transwell实验和Transwell侵袭小室实验检测2组SMMC-7721肝癌细胞的增殖、凋亡、转移和侵袭能力, 采用t检验对两组间数据进行统计学分析。 结果 NCEH1基因在肝癌组织中的平均表达量高于癌旁组织(本院标本Z=2.263, P=0.024, ICGC数据库U=18 768, P<0.001)。NCEH1基因在中等侵袭转移潜能的SMMC-7721细胞系中的表达量最高; 在低侵袭转移潜能的Bel-7402和HepG2细胞系中的表达水平次之, 在无侵袭转移潜能的Hep3B细胞系中的表达水平最低。KD组SMMC-7721细胞中NCEH1基因的表达水平明显低于NC组(t=11.578, P=0.000 3), NCEH1基因的敲减效率高达74.0%。较之NC组, KD组细胞生长速度明显减缓(t=32.10, P<0.001);细胞凋亡率明显升高(t=27.303, P<0.001);迁移率、转移和侵袭细胞数均明显降低(t值分别为9.51、38.123、22.331, P值均<0.001)。 结论 NCEH1基因在肝癌组织及细胞系中的表达明显升高, 且可促进肝癌细胞的生长增殖及侵袭转移并抑制凋亡, 提示其可能是一个潜在的肝癌治疗靶点。 -
关键词:
- 癌, 肝细胞 /
- 中性胆固醇酯水解酶1 /
- RNA, 小分子干扰
Abstract:Objective To investigate the expression of the neutral cholesterol ester hydrolase 1 (NCEH1) gene in liver cancer tissue and human hepatoma cell lines and the effect of NCEH1 gene knockdown on the proliferation, apoptosis, invasion, and metastasis abilities of human hepatoma SMMC-7721 cells. Methods Liver cancer tissue samples and adjacent tissue samples were collected from 32 patients with liver cancer who underwent surgical treatment in Guangzhou Red Cross Hospital Affiliated to Jinan University from January 2013 to June 2019, and quantitative real-time PCR was used to measure the relative expression level of the NCEH1 gene. Gene expression data of liver cancer samples up to September 2020 were downloaded from the ICGC database, and R software was used to analyze the data and obtain the expression level of the NCEH1 gene in each sample. The paired Wilcoxon signed-rank test and the Wilcoxon rank-sum test were used to investigate the differences between liver cancer tissue and adjacent tissue. Quantitative real-time PCR was used to measure the expression level of the NCEH1 gene in human hepatoma SMMC-7721, Bel-7402, HepG2, and Hep3B cells and normal human HL7702 liver cells. The lentivirus-mediated small interfering RNA (siRNA) technique was used to establish a human hepatoma SMMC-7721 cell line with NCEH1 gene knockdown, and the cells were divided into NCEH1 knockdown group (KD group) and negative control group (NC group); quantitative real-time PCR was used to measure the knockdown efficiency of the NCEH1 gene, and then MTT assay, flow cytometry with Annexin V-APC single staining, wound healing assay, Transwell assay, and Transwell chamber invasion assay were used to measure the proliferation, apoptosis, metastasis, and invasion abilities of SMMC-7721 cells in both groups. The t-test was used for statistical analysis of data between the two groups. Results The mean expression level of the NCEH1 gene in liver cancer tissue was significantly higher than that in adjacent tissue (specimens from our hospital: Z=2.263, P=0.024; ICGC database: U=18 768, P < 0.001). SMMC-7721 cell line with moderate potential of invasion and metastasis had the highest expression level of the NCEH1 gene, followed by BEL-7402 and HepG2 cell lines with low potential of invasion and metastasis, and Hep3B cell line without the potential of invasion and metastasis had the lowest expression level. The KD group had a significantly lower expression level of the NCEH1 gene than the NC group (t=11.578, P=0.000 3), and the knockdown efficiency of the NCEH1 gene was as high as 74.0%. Compared with the NC group, the KD group had a significant reduction in cell growth rate, a significant increase in apoptosis rate, and significant reductions in migration rate and the number of metastatic and invasive cells (t=32.100, 27.303, 9.51, 38.123, and 22.331, all P < 0.001). Conclusion There is a significant increase in the expression of the NCEH1 gene in liver cancer tissue and cell lines, and the NCEH1 gene can promote the growth, proliferation, invasion, and metastasis of hepatoma cells and inhibit their apoptosis, suggesting that it may be a potential therapeutic target for liver cancer. -
-
[1] CHEN H, JIA W. Progress in hepatectomy for hepatocellular carcinoma and peri-operation management[J]. Genes Dis, 2020, 7(3): 320-327. DOI: 10.1016/j.gendis.2020.02.001. [2] ALI ES, RYCHKOV GY, BARRITT GJ. Deranged hepatocyte intracellular Ca2+ homeostasis and the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma[J]. Cell Calcium, 2019, 82: 102057. DOI: 10.1016/j.ceca.2019.102057. [3] ALY SM, FETAIH HA, HASSANIN A, et al. Protective effects of garlic and cinnamon oils on hepatocellular carcinoma in albino rats[J]. Anal Cell Pathol (Amst), 2019, 2019: 9895485. DOI: 10.1155/2019/9895485. [4] IGARASHI M, OSUGA J, UOZAKI H, et al. The critical role of neutral cholesterol ester hydrolase 1 in cholesterol removal from human macrophages[J]. Circ Res, 2010, 107(11): 1387-1395. DOI: 10.1161/CIRCRESAHA.110.226613. [5] CHANG JW, NOMURA DK, CRAVATT BF. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis[J]. Chem Biol, 2011, 18(4): 476-484. DOI: 10.1016/j.chembiol.2011.02.008. [6] SHI WJ, LI XC, WEN HX, et al. Bioinformatics analysis of AL360181.1 regulating the progression and prognosis of colorectal cancer[J]. J Xi'an Univ (Med Sci), 2020, 41(5): 731-737. DOI: 10.7652/jdyxb202005018.史维俊, 李欣灿, 温贺新, 等. 基因AL360181.1调节结直肠癌进展及预后的生物信息学分析[J]. 西安交通大学学报(医学版), 2020, 41(5): 731-737. DOI: 10.7652/jdyxb202005018. [7] YE Z, WANG S, CHEN W, et al. Fat mass and obesity-associated protein promotes the tumorigenesis and development of liver cancer[J]. Oncol Lett, 2020, 20(2): 1409-1417. DOI: 10.3892/ol.2020.11673. [8] LI P, LIU Y. Effect of hepatitis B x gene on the expression of major histocompatibility complex class Ⅰ chain-related gene A-A5.1, invasion, and migration of HepG2.2.15 cells[J]. J Clin Hepatol, 2020, 36(4): 808-812. DOI: 10.3969/j.issn.1001-5256.2020.04.020.李沛, 刘宇. HBx基因对HepG2.2.15细胞MICA-A5.1表达及侵袭、迁移的影响[J]. 临床肝胆病杂志, 2020, 36(4): 808-812. DOI: 10.3969/j.issn.1001-5256.2020.04.020. [9] HO SY, HSU CY, LIU PH, et al. Albumin-bilirubin (ALBI) grade-based nomogram to predict tumor recurrence in patients with hepatocellular carcinoma[J]. Eur J Surg Oncol, 2019, 45(5): 776-781. DOI: 10.1016/j.ejso.2018.10.541. [10] YUAN SX, ZHOU WP. Progress and hot spots of comprehensive treatment for primary liver cancer[J]. Chin J Dig Surg, 2021, 20(2): 163-170. DOI: 10.3760/cma.j.cn115610-20201211-00776.袁声贤, 周伟平. 原发性肝癌综合治疗的进展和热点[J]. 中华消化外科杂志, 2021, 20(2): 163-170. DOI: 10.3760/cma.j.cn115610-20201211-00776. [11] JIANG Y, SUN A, ZHAO Y, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma[J]. Nature, 2019, 567(7747): 257-261. DOI: 10.1038/s41586-019-0987-8. [12] LIN L, DING Y, WANG Y, et al. Functional lipidomics: Palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism[J]. Hepatology, 2017, 66(2): 432-448. DOI: 10.1002/hep.29033. [13] POPE ED 3rd, KIMBROUGH EO, VEMIREDDY LP, et al. Aberrant lipid metabolism as a therapeutic target in liver cancer[J]. Expert Opin Ther Targets, 2019, 23(6): 473-483. DOI: 10.1080/14728222.2019.1615883. [14] NAKAGAWA H, HAYATA Y, KAWAMURA S, et al. Lipid metabolic reprogramming in hepatocellular carcinoma[J]. Cancers (Basel), 2018, 10(11): 447. DOI: 10.3390/cancers10110447. [15] CHISTIAKOV DA, BOBRYSHEV YV, OREKHOV AN. Macrophage-mediated cholesterol handling in atherosclerosis[J]. J Cell Mol Med, 2016, 20(1): 17-28. DOI: 10.1111/jcmm.12689. [16] LUCAS EK, DOUGHERTY SE, MCMEEKIN LJ, et al. PGC-1α provides a transcriptional framework for synchronous neurotransmitter release from parvalbumin-positive interneurons[J]. J Neurosci, 2014, 34(43): 14375-14387. DOI: 10.1523/JNEUROSCI.1222-14.2014. [17] CHIANG KP, NIESSEN S, SAGHATELIAN A, et al. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling[J]. Chem Biol, 2006, 13(10): 1041-1050. DOI: 10.1016/j.chembiol.2006.08.008. [18] NOMURA DK, FUJIOKA K, ISSA RS, et al. Dual roles of brain serine hydrolase KIAA1363 in ether lipid metabolism and organophosphate detoxification[J]. Toxicol Appl Pharmacol, 2008, 228(1): 42-48. DOI: 10.1016/j.taap.2007.11.021. [19] CHEUNG L, FISHER RM, KUZMINA N, et al. Psoriasis skin inflammation-induced microRNA-26b targets NCEH1 in underlying subcutaneous adipose tissue[J]. J Invest Dermatol, 2016, 136(3): 640-648. DOI: 10.1016/j.jid.2015.12.008. [20] ZHOU A, PAN H, SUN D, et al. miR-26b-5p inhibits the proliferation, migration and invasion of human papillary thyroid cancer in a β-catenin-dependent manner[J]. Onco Targets Ther, 2020, 13: 1593-1603. DOI: 10.2147/OTT.S236319. [21] WANG Y, SUN B, ZHAO X, et al. Twist1-related miR-26b-5p suppresses epithelial-mesenchymal transition, migration and invasion by targeting SMAD1 in hepatocellular carcinoma[J]. Oncotarget, 2016, 7(17): 24383-24401. DOI: 10.18632/oncotarget.8328. [22] KHOSLA R, HEMATI H, RASTOGI A, et al. miR-26b-5p helps in EpCAM+cancer stem cells maintenance via HSC71/HSPA8 and augments malignant features in HCC[J]. Liver Int, 2019, 39(9): 1692-1703. DOI: 10.1111/liv.14188. [23] GUPTA M, CHANDAN K, SARWAT M. Role of microRNA and long non-coding RNA in hepatocellular carcinoma[J]. Curr Pharm Des, 2020, 26(4): 415-428. DOI: 10.2174/1381612826666200115093835. -



 PDF下载 ( 3035 KB)
PDF下载 ( 3035 KB)

 下载:
下载: