miR-152-3p靶蛋白差异表达及调控机制在肝癌复发中的作用
DOI: 10.3969/j.issn.1001-5256.2021.02.023
Role of differential expression and regulatory mechanism of miR-152-3p target proteins in the recurrence of hepatocellular carcinoma
-
摘要:
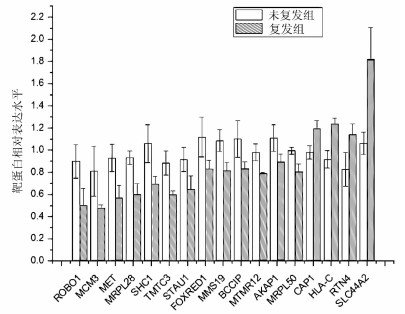
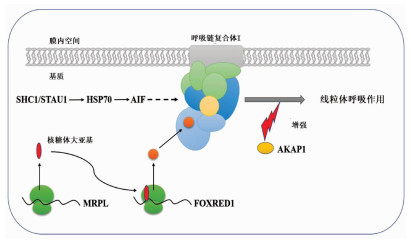
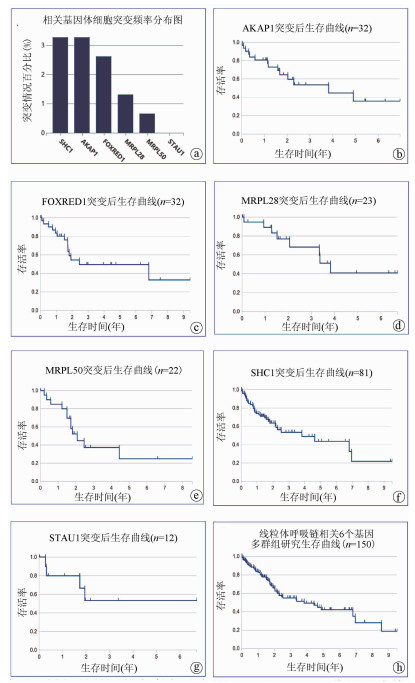
目的 比较肝癌复发与预后良好患者癌组织的蛋白表达,分析miR-152-3p相关的靶蛋白差异及调控机制,探讨miR-152-3p在肝癌复发中的作用。 方法 应用TMT标记全蛋白质组学测序方法和RT-PCR分别检测肝癌切除术后2年复发(n=6)与5年预后良好(n=6)患者肝癌组织的蛋白质表达和miR-152-3p的表达水平;联合六大数据库检索分析miR-152-3p靶基因,并运用Gene Ontology、DAVID和REACTOME数据库进行靶基因筛选、富集注释和信号转导通路富集分析;将筛选的miR-152-3p靶基因进行基因突变频率和生存曲线分析,验证miR-152-3p靶基因在肝癌复发中的作用。计量资料2组间比较采用独立样本t检验。肝脏组织有关基因生存率分析采用Kaplan-Meier分析。 结果 肝癌切除术后预后良好患者癌组织的miR-152-3p转录表达水平显著高于复发患者癌组织的水平(P<0.05)。蛋白测序结果显示,预后良好患者与复发患者的癌组织存在365个差异表达蛋白,联合肝癌复发数据库分析显示,其中有17个蛋白受miR-152-3p调控。进一步的信号通路分析发现,17个受miR-152-3p调控的靶基因其功能集中于线粒体核糖体翻译调控;多重富集发现靶基因与线粒体呼吸链复合体密切关系的蛋白6个,分别为AKAP1、FOXRED1、MRPL28、MRPL50、SHC1、STAU1。基因突变频率及生存曲线分析发现,线粒体呼吸链相关靶蛋白的功能缺失或减弱会严重影响患者的生存率。 结论 miR-152-3p在HCC切除术后预后良好和复发患者的癌组织表达差异显著,miR-152-3p在肝癌复发中作用机制可能是通过调控靶基因AKAP1、FOXRED1、MRPL28、MRPL50、SHC1、STAU1作用于线粒体呼吸链,影响细胞氧化呼吸功能导致肝癌复发。 -
关键词:
- 癌, 肝细胞 /
- miR-152-3p /
- 复发
Abstract:Objective To investigate the difference in protein expression between hepatocellular carcinoma (HCC) patients with recurrence and those with good prognosis, the differential expression and regulatory mechanism of miR-152-3p target proteins, and the role of miR-152-3p in the recurrence of HCC. Methods TMT-labeled proteomic sequencing and RT-PCR were used to measure the expression of proteins and the expression of miR-152-3p in the HCC tissue of six patients with recurrence at 2 years after HCC resection and six patients with good prognosis at 5 years. Six databases were used to analyze the target genes of miR-152-3p, and Gene Ontology, DAVID, and REACTOME databases were used to perform target gene screening, enrichment annotation, and signal transduction pathway enrichment analysis. Gene mutation frequency and survival curve analysis were performed for the target genes of miR-152-3p to verify the role of miR-152-3p target genes in patients with HCC recurrence. The independent samples t-test was used for comparison of continuous data between two groups, and a Kaplan-Meier analysis was performed to investigate the survival rates of liver-related genes. Results Compared with the patients with HCC recurrence, the patients with good prognosis after HCC resection had a significantly higher transcriptional expression level of miR-152-3p in HCC tissue (P < 0.05). The results of protein sequencing showed that there were 365 differentially expressed proteins in HCC tissue between the patients with good prognosis and the patients with recurrence, and the analysis of HCC recurrence databases showed that 17 proteins were regulated by miR-152-3p. Further analysis of the signaling pathways showed that the function of the 17 target genes regulated by miR-152-3p was enriched in the translation and regulation of mitochondria and ribosome, and multiple enrichment revealed that six target genes were closely associated with mitochondrial respiratory chain complex, i.e., AKAP1, FOXRED1, MRPL28, MRPL50, SHC1, and STAU1. Gene mutation frequency and survival curve analysis showed that the loss or weakening of the function of mitochondrial respiratory chain-related target proteins seriously affected the prognosis and survival rate of patients. Conclusion There is a significant difference in the expression of miR-152-3p in HCC tissue between patients with good prognosis and those with recurrence after HCC resection, and miR-152-3p may lead to the recurrence of HCC by regulating the target genes AKAP1, FOXRED1, MRPL28, MRPL50, SHC1, and STAU1, acting on the mitochondrial respiratory chain, and affecting the oxidative respiratory function of cells. -
Key words:
- Carcinoma, Hepatocellular /
- miR-152-3p /
- Recurrence
-
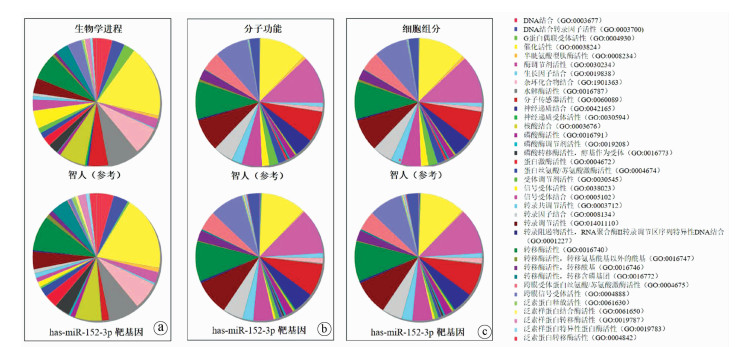
表 1 miR-152-3p靶蛋白的GO富集分析结果
GO编号 项目 P值 Gene ID 生物学过程 GO: 0070125 线粒体翻译延伸 7.30×10-3 ENSG00000086504, ENSG00000136897 GO: 0070126 线粒体翻译终止 7.40×10-3 ENSG00000086504, ENSG00000136897 GO: 0007165 信号转导 8.03×10-3 ENSG00000131236, ENSG00000160691… 细胞成分 GO: 0005762 线粒体大核糖体亚基 4.13×10-3 ENSG00000086504, ENSG00000136897 GO: 0005886 质膜 4.93×10-3 ENSG00000124214, ENSG00000169855… GO: 0005743 细胞线粒体内膜 5.60×10-3 ENSG00000110074, ENSG00000086504… GO: 0009986 细胞表面 8.05×10-3 ENSG00000169855, ENSG00000204525… 分子功能 GO: 0044822 Poly(A)RNA结合 1.49×10-3 ENSG00000074527, ENSG00000156587… GO: 0005515 蛋白质结合 8.02×10-3 ENSG00000215301, ENSG00000100077… 表 2 miR-152-3p靶蛋白的相关信号通路富集分析结果
REACTOME编号 项目 P值 Gene ID R-HSA-428890 Slit/Robo信号通路 1.08×10-3 ENSG00000169855, ENSG00000131236 R-HSA-5419276 线粒体翻译终止 9.94×10-3 ENSG00000086504, ENSG00000136897 R-HSA-5368286 线粒体翻译启示 9.94×10-3 ENSG00000086504, ENSG00000136897 R-HSA-5389840 线粒体翻译延伸 9.94×10-3 ENSG00000086504, ENSG00000136897 表 3 miR-152-3p靶基因的相关疾病富集分析结果
编号 项目 P值 Gene ID 1 蛋白质定量性状位点 4.99×10-3 ENSG00000169855,
ENSG000002045252 获得性免疫缺陷综合征 5.78×10-3 ENSG00000110074,
ENSG00000086504…3 头颈癌 8.75×10-3 ENSG00000204525,
ENSG000001059764 乳腺癌 9.28×10-3 ENSG00000160691,
ENSG00000107949…表 4 miR-152-3p靶蛋白的组织定位富集分析结果
组织库编号 项目 P值 Gene ID 26802 正常胎盘组织 0.91×10-4 ENSG00000160691,
ENSG00000107949…21061 正常结肠组织 5.68×10-3 ENSG00000169855,
ENSG00000129353…899 正常脾组织 1.27×10-2 ENSG00000131236,
ENSG00000204525…表 5 miR-152-3p作用于线粒体呼吸链相关基因的突变频率统计表
基因名 组内体细胞突变频率 GDC数据库内体细胞突变频率 拷贝数增加 拷贝数减少 SHC1 5/150(3.33%) 163/10 202 74/152(48.68%) 4/152(2.63%) AKAP1 5/150(3.33%) 182/10 202 26/152(17.11%) 3/152(1.97%) FOXRED1 4/150(2.67%) 112/10 202 3/152(1.97%) 25/152(16.45%) MRPL28 2/150(1.33%) 58/10 202 9/152(5.92%) 12/152(7.89%) MRPL50 1/150(0.67%) 63/10 202 9/152(5.92%) 13/152(8.55%) STAU1 0/150(0.00%) 147/10 202 9/152(5.92%) 4/152(2.63%) -
[1] DUTTA R, MAHATO RI. Recent advances in hepatocellular carcinoma therapy[J]. Pharmacol Ther, 2017, 173(5): 106-117. [2] ZHANG Z, MA L, GOSWAMI S, et al. Landscape of infiltrating B cells and their clinical significance in human hepatocellular carcinoma[J]. OncoImmunology, 2019, 8(4): 1-11. [3] ZHAO ZJ, HU Y, SHEN XH, et al. HBx represses RIZ1 expression by DNA methyltransferase 1 involvement in decreased miR-152 in hepatocellular carcinoma[J]. Oncol Rep, 2017, 37(5): 2811. DOI: 10.3892/or.2017.5518 [4] DUAN YF, ZHAO QX, JING X. Effect of long non-coding RNAs on glycolytic pathway in primary liver cancer and related mechanisms[J]. J Clin Hepatol, 2019, 35(6): 1374-1376. (in Chinese) DOI: 10.3969/j.issn.1001-5256.2019.06.043段艺菲, 赵清喜, 荆雪.长链非编码RNA对原发性肝癌中糖酵解途径的影响机制[J].临床肝胆病杂志, 2019, 35(6): 1374-1376. DOI: 10.3969/j.issn.1001-5256.2019.06.043 [5] DING J, DUAN BW, DI L. Expression and mechanism of microRNA and Na+/H+ exchanger regulatory factor 1 on hepatocellular carcinoma[J/CD]. Chin J Liver Dis (Electronic Version), 2020, 12(3): 59-64.(in Chinese) https://www.cnki.com.cn/Article/CJFDTOTAL-GZBZ202003015.htm丁兢, 段斌炜, 邸亮.微小RNA-320a和钠氢交换调控因子1在肝细胞癌中的表达及机制[J/CD].中国肝脏病杂志(电子版), 2020, 12(3): 59-64. https://www.cnki.com.cn/Article/CJFDTOTAL-GZBZ202003015.htm [6] QU ZN, LIN L, ZHONG W, et al. Meta-analysis of value of circulating microRNA 122 on diagnosis of hepatocellular carcinoma[J/CD]. Chin J Liver Dis (Electronic Version), 2020, 12(3): 23-28.(in Chinese) https://www.cnki.com.cn/Article/CJFDTOTAL-GZBZ202003007.htm屈振南, 林霖, 钟伟, 等.循环微小RNA-122诊断肝细胞癌的Meta分析[J/CD].中国肝脏病杂志(电子版), 2020, 12(3): 23-28. https://www.cnki.com.cn/Article/CJFDTOTAL-GZBZ202003007.htm [7] ZHOU JJ, ZHANG YJ, QI YH, et al. MicroRNA-152 inhibits tumor cell growth by directly targeting RTKN in hepatocellular carcinoma[J]. Oncol Rep, 2017, 37(2): 1227. DOI: 10.3892/or.2016.5290 [8] MIQUELESTORENA-STANDLEY E, TALLET A, COLLIN C, et al. Interest of variations in microRNA-152 and-122 in a series of hepatocellular carcinomas related to hepatitis C virus infection[J]. Hepatol Res, 2018, 48(7): 566-573. DOI: 10.1111/hepr.12915 [9] BILIC′ P, GUILLEMIN N, KOVACˇEVIC′ A, et al. Serum proteome profiling in canine idiopathic dilated cardiomyopathy using TMT-based quantitative proteomics approach[J]. J Proteomics, 2018, 179: 110-121. DOI: 10.1016/j.jprot.2018.03.007 [10] CHANG K, JIANG ZY, REN JL, et al. Bioinformatics prediction of miR-130b-3p target genes associated with diabetic nephropathy[J]. J Chongqing Med Univ, 2018, 43(6): 788-792. (in Chinese) https://www.cnki.com.cn/Article/CJFDTOTAL-ZQYK201806011.htm常凯, 江忠勇, 任俊龙, 等.糖尿病肾病相关miR-130b-3p靶点的生物信息学预测[J].重庆医科大学学报, 2018, 43(6): 788-792. https://www.cnki.com.cn/Article/CJFDTOTAL-ZQYK201806011.htm [11] CHEN L, ZHANG YH, WANG SP, et al. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways[J]. PLoS One, 2017, 12(9): e0184129. DOI: 10.1371/journal.pone.0184129 [12] WANG F, YING H, HE B, et al. Circulating miR-148/152 family as potential biomarkers in hepatocellular carcinoma[J]. Tumour Biol, 2016, 37(4): 4945-4953. DOI: 10.1007/s13277-015-4340-z [13] HUANG J, WANG Y, GUO Y, et al. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1[J]. Hepatology, 2010, 52(1): 60-70. DOI: 10.1002/hep.23660 [14] DANG YW, ZENG J, HE RQ, et al. Effects of miR-152 on cell growth inhibition, motility suppression and apoptosis induction in hepatocellular carcinoma cells[J]. Asian Pac J Cancer Prev, 2014, 15(12): 4969-4976. DOI: 10.7314/APJCP.2014.15.12.4969 [15] IRYNA K, VOLODYMYR T, ALINE DC, et al. MicroRNA-152-mediated dysregulation of hepatic transferrin receptor 1 in liver carcinogenesis[J]. Oncotarget, 2016, 7(2): 1276-1287. DOI: 10.18632/oncotarget.6004 [16] FORMOSA LE, MASAKAZU M, FRAZIER AE, et al. Characterization of mitochondrial FOXRED1 in the assembly of respiratory chain complex I[J]. Hum Mol Genet, 2015, 24(10): 2952-2965. DOI: 10.1093/hmg/ddv058 [17] DAGDA RK, GUSDON AM, PIEN I, et al. Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson's disease[J]. Cell Death Differ, 2011, 18(12): 1914-1923. DOI: 10.1038/cdd.2011.74 [18] YU J, ZHANG Y, ZHOU DX, et al. Higher expression of A-kinase anchoring-protein 1 predicts poor prognosis in human hepatocellular carcinoma[J]. Oncol Lett, 2018, 16(1): 131-136. [19] JIANG ZJ, CHEN J, LI LQ. Research progress of long non-coding RNA in the invasion and metastasis of hepatocellular carcinoma[J]. J Med Postgraduates, 2019, 32(7): 765-770. (in Chinese) https://www.cnki.com.cn/Article/CJFDTOTAL-JLYB201907020.htm蒋志俊, 陈洁, 黎乐群.长链非编码RNA在肝癌侵袭转移中的研究进展[J].医学研究生学报, 2019, 32(7): 765-770. https://www.cnki.com.cn/Article/CJFDTOTAL-JLYB201907020.htm [20] ZHANG XC, LI B. Towards understanding the mechanisms of proton pumps in Complex-Ⅰ of the respiratory chain[J]. Biophy Rep, 2019, 5(5-6): 219-234. DOI: 10.1007/s41048-019-00094-7 -



 PDF下载 ( 4086 KB)
PDF下载 ( 4086 KB)

 下载:
下载: