| [1] |
YOUNOSSI ZM, KOENIG AB, ABDELATIF D, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes[J]. Hepatology, 2016, 64( 1): 73- 84. DOI: 10.1002/hep.28431. |
| [2] |
LI J, LI YN, YANG JB, et al. Application of single cell sequencing technology in liver regeneration[J]. Chin J Dig Surg, 2023, 22( 5): 663- 666. DOI: 10.3760/cma.j.cn115610-20230316-00109. |
| [3] |
HUNDERTMARK J, BERGER H, TACKE F. Single cell RNA sequencing in NASH[J]. Methods Mol Biol, 2022, 2455: 181- 202. DOI: 10.1007/978-1-0716-2128-8_15. |
| [4] |
SU Q, KIM SY, ADEWALE F, et al. Single-cell RNA transcriptome landscape of hepatocytes and non-parenchymal cells in healthy and NAFLD mouse liver[J]. iScience, 2021, 24( 11): 103233. DOI: 10.1016/j.isci.2021.103233. |
| [5] |
KODA Y, TERATANI T, CHU PS, et al. CD8 + tissue-resident memory T cells promote liver fibrosis resolution by inducing apoptosis of hepatic stellate cells[J]. Nat Commun, 2021, 12( 1): 4474. DOI: 10.1038/s41467-021-24734-0. |
| [6] |
LI WG, ZHOU YW, ZHANG XZ. Level and correlation of peripheral blood hypoxia-inducible factor-1α, regulatory T cell and helper T cell and inflammatory cytokines in patients with non-alcoholic fatty liver disease[J/CD]. Chin J Liver Dis Electron Version, 2020, 12( 4): 83- 88. DOI: 10.3969/j.issn.1674-7380.2020.04.015. |
| [7] |
LI Y, DU ZX, WANG MT, et al. Correlation between disease progression and levels of T cells in patients with non-alcoholic fatty liver disease[J]. J Clin Med Pract, 2020, 24( 14): 11- 14, 18. DOI: 10.7619/jcmp.202014003. 李阳, 杜志祥, 王牧婷, 等. 非酒精性脂肪性肝病患者病情进展与T细胞水平的相关性研究[J]. 实用临床医药杂志, 2020, 24( 14): 11- 14, 18. DOI: 10.7619/jcmp.202014003. |
| [8] |
LI HY, GAO YX, WU JC, et al. Single-cell transcriptome reveals a novel mechanism of C-Kit +-liver sinusoidal endothelial cells in NASH[J]. Cell Biosci, 2024, 14( 1): 31. DOI: 10.1186/s13578-024-01215-7. |
| [9] |
National Workshop on Fatty Liver and Alcoholic Liver Disease, Chinese Society of Hepatology, Chinese Medical Association, Fatty Liver Expert Committee, Chinese Medical Doctor Association. Guidelines of prevention and treatment for nonalcoholic fatty liver disease: A 2018 update[J]. J Clin Hepatol, 2018, 34( 5): 947- 957. DOI: 10.3969/j.issn.1001-5256.2018.05.007. |
| [10] |
YOUNOSSI Z, TACKE F, ARRESE M, et al. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis[J]. Hepatology, 2019, 69( 6): 2672- 2682. DOI: 10.1002/hep.30251. |
| [11] |
ZHANG SL, CAO MM, YANG F, et al. Analysis of the change trend of etiological burden of disease of liver cancer in the Chinese population from 1990 to 2019[J]. Chin J Dig Surg, 2023, 22( 1): 122- 130. DOI: 10.3760/cma.j.cn115610-20221112-00687. |
| [12] |
HARDY T, OAKLEY F, ANSTEE QM, et al. Nonalcoholic fatty liver disease: Pathogenesis and disease spectrum[J]. Annu Rev Pathol, 2016, 11: 451- 496. DOI: 10.1146/annurev-pathol-012615-044224. |
| [13] |
NOUREDDIN M, SANYAL AJ. Pathogenesis of NASH: The impact of multiple pathways[J]. Curr Hepatol Rep, 2018, 17( 4): 350- 360. DOI: 10.1007/s11901-018-0425-7. |
| [14] |
WOLF MJ, ADILI A, PIOTROWITZ K, et al. Metabolic activation of intrahepatic CD8 + T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes[J]. Cancer Cell, 2014, 26( 4): 549- 564. DOI: 10.1016/j.ccell.2014.09.003. |
| [15] |
HER Z, TAN JHL, LIM YS, et al. CD4 + T cells mediate the development of liver fibrosis in high fat diet-induced NAFLD in humanized mice[J]. Front Immunol, 2020, 11: 580968. DOI: 10.3389/fimmu.2020.580968. |
| [16] |
|
| [17] |
LI DR, ZHANG ZJ, ZHANG C, et al. Unraveling the connection between Hashimoto’s Thyroiditis and non-alcoholic fatty liver disease: Exploring the role of CD4 + central memory T cells through integrated genetic approaches[J]. Endocrine, 2024, 85( 2): 751- 765. DOI: 10.1007/s12020-024-03745-z. |
| [18] |
HUANG YY, LIU X, WANG HY, et al. Single-cell transcriptome landscape of zebrafish liver reveals hepatocytes and immune cell interactions in understanding nonalcoholic fatty liver disease[J]. Fish Shellfish Immunol, 2024, 146: 109428. DOI: 10.1016/j.fsi.2024.109428. |
| [19] |
YU Q, SHARMA A, OH SY, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma[J]. Nat Immunol, 2009, 10( 9): 992- 999. DOI: 10.1038/ni.1762. |
| [20] |
ROLLA S, ALCHERA E, IMARISIO C, et al. The balance between IL-17 and IL-22 produced by liver-infiltrating T-helper cells critically controls NASH development in mice[J]. Clin Sci(Lond), 2016, 130( 3): 193- 203. DOI: 10.1042/CS20150405. |
| [21] |
WANG ZY. Review on the mechanism of T-cell factor 3 in the progression and metastasis of hepatocellular carcinoma[J]. J World Latest Med Inf, 2024, 24( 16): 43- 46. DOI: 10.3969/j.issn.1671-3141.2024.016.009. |
| [22] |
GUI MH, HUANG SL, LI SZ, et al. Integrative single-cell transcriptomic analyses reveal the cellular ontological and functional heterogeneities of primary and metastatic liver tumors[J]. J Transl Med, 2024, 22( 1): 206. DOI: 10.1186/s12967-024-04947-9. |
| [23] |
UNUTMAZ D, XIANG W, SUNSHINE MJ, et al. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse[J]. J Immunol, 2000, 165( 6): 3284- 3292. DOI: 10.4049/jimmunol.165.6.3284. |
| [24] |
DUDEK M, PFISTER D, DONAKONDA S, et al. Auto-aggressive CXCR6 + CD8 T cells cause liver immune pathology in NASH[J]. Nature, 2021, 592( 7854): 444- 449. DOI: 10.1038/s41586-021-03233-8. |
| [25] |
WEHR A, BAECK C, HEYMANN F, et al. Chemokine receptor CXCR6-dependent hepatic NK T Cell accumulation promotes inflammation and liver fibrosis[J]. J Immunol, 2013, 190( 10): 5226- 5236. DOI: 10.4049/jimmunol.1202909. |



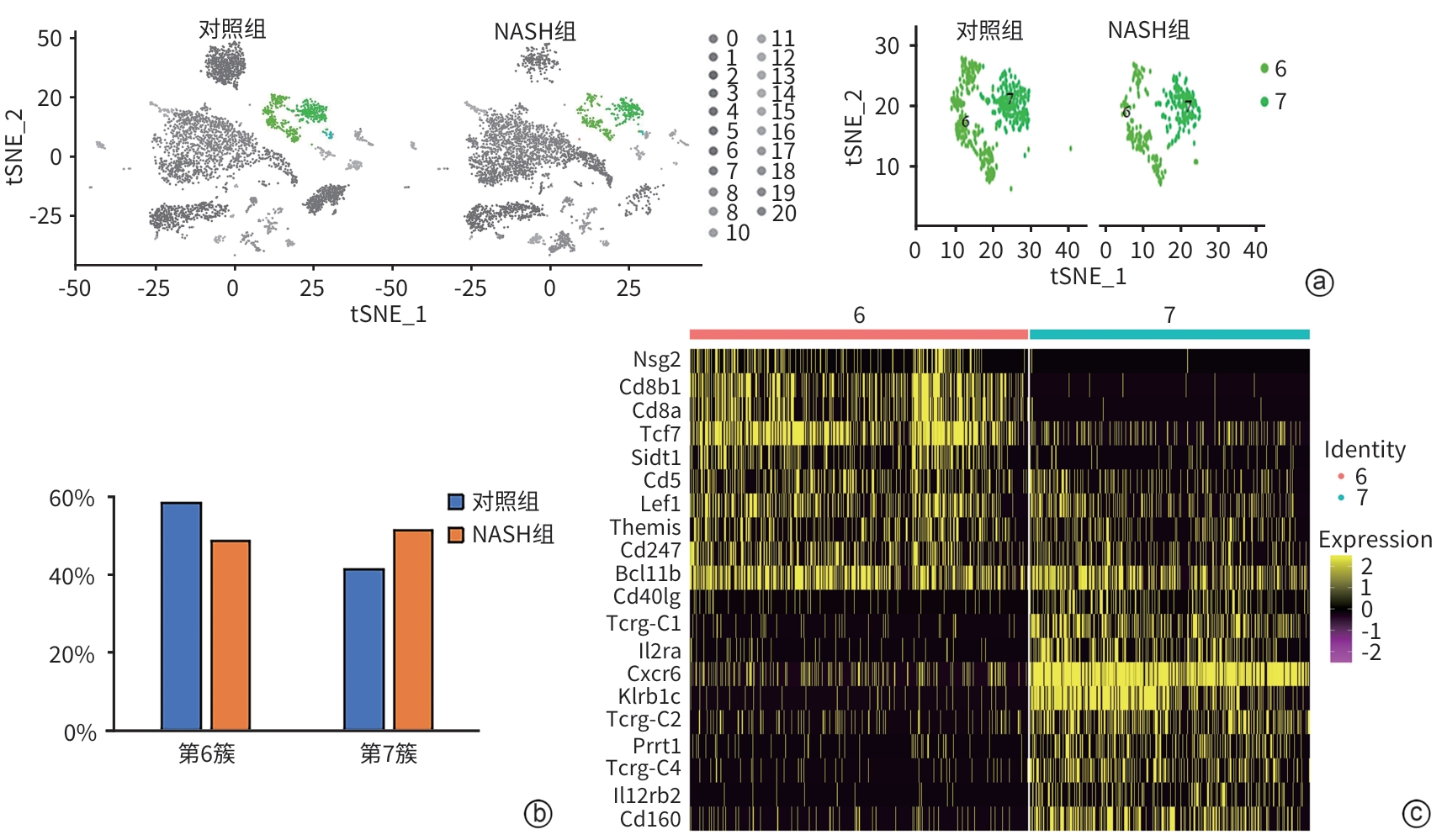




 DownLoad:
DownLoad: