| [1] |
YOUNOSSI ZM, GOLABI P, PAIK JM, et al. The global epidemiology of nonalcoholic fatty liver disease(NAFLD) and nonalcoholic steatohepatitis(NASH): a systematic review[J]. Hepatology, 2023, 77( 4): 1335- 1347. DOI: 10.1097/HEP.0000000000000004. |
| [2] |
SARIN SK, KUMAR M, ESLAM M, et al. Liver diseases in the Asia-Pacific region: a Lancet Gastroenterology& Hepatology Commission[J]. Lancet Gastroenterol Hepatol, 2020, 5( 2): 167- 228. DOI: 10.1016/S2468-1253(19)30342-5. |
| [3] |
ANDROUTSAKOS T, NASIRI-ANSARI N, BAKASIS AD, et al. SGLT-2 inhibitors in NAFLD: expanding their role beyond diabetes and cardioprotection[J]. Int J Mol Sci, 2022, 23( 6). DOI: 10.3390/ijms23063107. |
| [4] |
KOU XX, ZHANG H, DENG JX, et al. Role of intrahepatic microenvironment induced-autophagy in nonalcoholic fatty liver disease[J]. J Clin Hepatol, 2023, 39( 6): 1440- 1445. DOI: 10.3969/j.issn.1001-5256.2023.06.029. |
| [5] |
|
| [6] |
FILALI-MOUNCEF Y, HUNTER C, ROCCIO F, et al. The ménage à trois of autophagy, lipid droplets and liver disease[J]. Autophagy, 2022, 18( 1): 50- 72. DOI: 10.1080/15548627.2021.1895658. |
| [7] |
KOCAK M, EZAZI ERDI S, JORBA G, et al. Targeting autophagy in disease: established and new strategies[J]. Autophagy, 2022, 18( 3): 473- 495. DOI: 10.1080/15548627.2021.1936359. |
| [8] |
YUAN C, LIAN QH, NI BB, et al. Screening and bioinformatics analysis of key autophagy-related genes in alcoholic hepatitis[J]. Ogran Transplant, 2024, 15( 1): 90- 101. DOI: 10.3969/j.issn.1674-7445.2023163. |
| [9] |
KIRCHNER P, BOURDENX M, MADRIGAL-MATUTE J, et al. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs[J]. PLoS Biol, 2019, 17( 5): e3000301. DOI: 10.1371/journal.pbio.3000301. |
| [10] |
SAHU R, KAUSHIK S, CLEMENT CC, et al. Microautophagy of cytosolic proteins by late endosomes[J]. Dev Cell, 2011, 20( 1): 131- 139. DOI: 10.1016/j.devcel.2010.12.003. |
| [11] |
WANG L, KLIONSKY DJ, SHEN HM. The emerging mechanisms and functions of microautophagy[J]. Nat Rev Mol Cell Biol, 2023, 24( 3): 186- 203. DOI: 10.1038/s41580-022-00529-z. |
| [12] |
OLZMANN JA, CARVALHO P. Dynamics and functions of lipid droplets[J]. Nat Rev Mol Cell Biol, 2019, 20( 3): 137- 155. DOI: 10.1038/s41580-018-0085-z. |
| [13] |
CHUNG J, PARK J, LAI ZW, et al. The Troyer syndrome protein spartin mediates selective autophagy of lipid droplets[J]. Nat Cell Biol, 2023, 25( 8): 1101- 1110. DOI: 10.1038/s41556-023-01178-w. |
| [14] |
BACKE SJ, SAGER RA, HERITZ JA, et al. Activation of autophagy depends on Atg1/Ulk1-mediated phosphorylation and inhibition of the Hsp90 chaperone machinery[J]. Cell Rep, 2023, 42( 7): 112807. DOI: 10.1016/j.celrep.2023.112807. |
| [15] |
XU Y, WANG S, LEUNG CK, et al. α-amanitin induces autophagy through AMPK-mTOR-ULK1 signaling pathway in hepatocytes[J]. Toxicol Lett, 2023, 383: 89- 97. DOI: 10.1016/j.toxlet.2023.06.004. |
| [16] |
BAI J, ZHU Y, HE L, et al. Saponins from bitter melon reduce lipid accumulation via induction of autophagy in C. elegans and HepG2 cell line[J]. Curr Res Food Sci, 2022, 5: 1167- 1175. DOI: 10.1016/j.crfs.2022.06.011. |
| [17] |
NGUYEN LN, BORMANN J, LE GT, et al. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection[J]. Fungal Genet Biol, 2011, 48( 3): 217- 224. DOI: 10.1016/j.fgb.2010.11.004. |
| [18] |
KURUSU T, KOYANO T, HANAMATA S, et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development[J]. Autophagy, 2014, 10( 5): 878- 888. DOI: 10.4161/auto.28279. |
| [19] |
de la BALLINA LR, MUNSON MJ, SIMONSEN A. Lipids and lipid-binding proteins in selective autophagy[J]. J Mol Biol, 2020, 432( 1): 135- 159. DOI: 10.1016/j.jmb.2019.05.051. |
| [20] |
BARROS J, MAGEN S, LAPIDOT-COHEN T, et al. Autophagy is required for lipid homeostasis during dark-induced senescence[J]. Plant Physiol, 2021, 185( 4): 1542- 1558. DOI: 10.1093/plphys/kiaa120. |
| [21] |
MALLÉN-PONCE MJ, GÁMEZ-ARCAS S, PÉREZ-PÉREZ ME. Redox partner interactions in the ATG8 lipidation system in microalgae[J]. Free Radic Biol Med, 2023, 203: 58- 68. DOI: 10.1016/j.freeradbiomed.2023.04.004. |
| [22] |
OUYANG Q, LIU R. MTOR-mediates hepatic lipid metabolism through an autophagic SNARE complex[J]. Autophagy, 2022, 18( 6): 1467- 1469. DOI: 10.1080/15548627.2022.2037853. |
| [23] |
ADNAN M, ISLAM W, ZHANG J, et al. Diverse role of SNARE protein Sec22 in vesicle trafficking, membrane fusion, and autophagy[J]. Cells, 2019, 8( 4): 337. DOI: 10.3390/cells8040337. |
| [24] |
SHROFF A, NAZARKO TY. SQSTM1, lipid droplets and current state of their lipophagy affairs[J]. Autophagy, 2023, 19( 2): 720- 723. DOI: 10.1080/15548627.2022.2094606. |
| [25] |
SCHULZE RJ, DRIŽYTĖ K, CASEY CA, et al. Hepatic lipophagy: new insights into autophagic catabolism of lipid droplets in the liver[J]. Hepatol Commun, 2017, 1( 5): 359- 369. DOI: 10.1002/hep4.1056. |
| [26] |
YANG M, LUO S, CHEN W, et al. Chaperone-mediated autophagy: a potential target for metabolic diseases[J]. Curr Med Chem, 2023, 30( 16): 1887- 1899. DOI: 10.2174/0929867329666220811141955. |
| [27] |
SEO AY, LAU PW, FELICIANO D, et al. AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation[J]. Elife, 2017, 6: e21690. DOI: 10.7554/eLife.21690. |
| [28] |
OKU M, MAEDA Y, KAGOHASHI Y, et al. Evidence for ESCRT- and clathrin-dependent microautophagy[J]. J Cell Biol, 2017, 216( 10): 3263- 3274. DOI: 10.1083/jcb.201611029. |
| [29] |
HOMMA Y, HIRAGI S, FUKUDA M. Rab family of small GTPases: an updated view on their regulation and functions[J]. FEBS J, 2021, 288( 1): 36- 55. DOI: 10.1111/febs.15453. |
| [30] |
LI Z, WELLER SG, DRIZYTE-MILLER K, et al. Maturation of lipophagic organelles in hepatocytes is dependent upon a Rab10/dynamin-2 complex[J]. Hepatology, 2020, 72( 2): 486- 502. DOI: 10.1002/hep.31059. |
| [31] |
DENG Y, ZHOU C, MIRZA AH, et al. Rab18 binds PLIN2 and ACSL3 to mediate lipid droplet dynamics[J]. Biochim Biophys Acta Mol Cell Biol Lipids, 2021, 1866( 7): 158923. DOI: 10.1016/j.bbalip.2021.158923. |
| [32] |
KLOSKA A, WĘSIERSKA M, MALINOWSKA M, et al. Lipophagy and lipolysis status in lipid storage and lipid metabolism diseases[J]. Int J Mol Sci, 2020, 21( 17). DOI: 10.3390/ijms21176113. |
| [33] |
SINGH R, KAUSHIK S, WANG Y, et al. Autophagy regulates lipid metabolism[J]. Nature, 2009, 458( 7242): 1131- 1135. DOI: 10.1038/nature07976. |
| [34] |
TAN YM, TAN YF, MENG GZ, et al. The regulatory role of lipophagy in lipid metabolism diseases[J]. J Med Sci Cent South China, 2022, 50( 5): 777- 780. DOI: 10.15972/j.cnki.43-1509/r.2022.05.039. |
| [35] |
CUI W, SATHYANARAYAN A, LOPRESTI M, et al. Lipophagy-derived fatty acids undergo extracellular efflux via lysosomal exocytosis[J]. Autophagy, 2021, 17( 3): 690- 705. DOI: 10.1080/15548627.2020.1728097. |
| [36] |
ZHAO N, TAN H, WANG L, et al. Palmitate induces fat accumulation via repressing FoxO1-mediated ATGL-dependent lipolysis in HepG2 hepatocytes[J]. PLoS One, 2021, 16( 1): e0243938. DOI: 10.1371/journal.pone.0243938. |
| [37] |
SATHYANARAYAN A, MASHEK MT, MASHEK DG. ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism[J]. Cell Rep, 2017, 19( 1): 1- 9. DOI: 10.1016/j.celrep.2017.03.026. |
| [38] |
ZHANG G, HAN J, WANG L, et al. The vesicular transporter STX11 governs ATGL-mediated hepatic lipolysis and lipophagy[J]. iScience, 2022, 25( 4): 104085. DOI: 10.1016/j.isci.2022.104085. |
| [39] |
LI L, LI Q, HUANG W, et al. Dapagliflozin alleviates hepatic steatosis by restoring autophagy via the AMPK-mTOR pathway[J]. Front Pharmacol, 2021, 12: 589273. DOI: 10.3389/fphar.2021.589273. |
| [40] |
ZHANG D, ZHANG Y, WANG Z, et al. Thymoquinone attenuates hepatic lipid accumulation by inducing autophagy via AMPK/mTOR/ULK1-dependent pathway in nonalcoholic fatty liver disease[J]. Phytother Res, 2023, 37( 3): 781- 797. DOI: 10.1002/ptr.7662. |
| [41] |
SEOK S, KIM YC, BYUN S, et al. Fasting-induced JMJD3 histone demethylase epigenetically activates mitochondrial fatty acid β-oxidation[J]. J Clin Invest, 2018, 128( 7): 3144- 3159. DOI: 10.1172/JCI97736. |
| [42] |
|
| [43] |
BYUN S, SEOK S, KIM YC, et al. Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase[J]. Nat Commun, 2020, 11( 1): 807. DOI: 10.1038/s41467-020-14384-z. |
| [44] |
ALSHEHADE S, ALSHAWSH MA, MURUGAIYAH V, et al. The role of protein kinases as key drivers of metabolic dysfunction-associated fatty liver disease progression: New insights and future directions[J]. Life Sci, 2022, 305: 120732. DOI: 10.1016/j.lfs.2022.120732. |
| [45] |
LIU YY, SUI M, JIANG XF, et al. Effect of Danzhi Tiaozhi decoction on the PI3K/AKT/FOXO1 signaling pathway in high-fat induced MAFLD rats[J]. J Nangjing Univ Tradit Chin Med, 2023, 39( 6): 541- 547. DOI: 10.14148/j.issn.1672-0482.2023.0541. |
| [46] |
WANG S, YANG FJ, SHANG LC, et al. Puerarin protects against high-fat high-sucrose diet-induced non-alcoholic fatty liver disease by modulating PARP-1/PI3K/AKT signaling pathway and facilitating mitochondrial homeostasis[J]. Phytother Res, 2019, 33( 9): 2347- 2359. DOI: 10.1002/ptr.6417. |
| [47] |
WANG MY, LI EW, GAO G, et al. Zexie Decoction regulates Akt/TFEB signaling pathway to promote lipophagy in hepatocytes[J]. China J Chin Mater Med, 2022, 47( 22): 6183- 6190. DOI: 10.19540/j.cnki.cjcmm.20220706.702. |
| [48] |
YAN H, CHAI CY, ZHANG D, et al. Explore the mechanism of autophagy and insulin resistance in non-alcoholic fatty liver disease based on JNK signaling pathway[J]. Shaanxi Med J, 2023, 52( 11): 1506- 1510. DOI: 10.3969/j.issn.1000-7377.2023.11.012. |
| [49] |
GONG J, GAO X, GE S, et al. The role of cGAS-STING signalling in metabolic diseases: from signalling networks to targeted intervention[J]. Int J Biol Sci, 2024, 20( 1): 152- 174. DOI: 10.7150/ijbs.84890. |
| [50] |
PANZITT K, WAGNER M. FXR in liver physiology: Multiple faces to regulate liver metabolism[J]. Biochim Biophys Acta Mol Basis Dis, 2021, 1867( 7): 166133. DOI: 10.1016/j.bbadis.2021.166133. |
| [51] |
MA SY, SUN KS, ZHANG M, et al. Disruption of Plin5 degradation by CMA causes lipid homeostasis imbalance in NAFLD[J]. Liver Int, 2020, 40( 10): 2427- 2438. DOI: 10.1111/liv.14492. |
| [52] |
YOU Y, LI WZ, ZHANG S, et al. SNX10 mediates alcohol-induced liver injury and steatosis by regulating the activation of chaperone-mediated autophagy[J]. J Hepatol, 2018, 69( 1): 129- 141. DOI: 10.1016/j.jhep.2018.01.038. |
| [53] |
LEE W, KIM HY, CHOI YJ, et al. SNX10-mediated degradation of LAMP2A by NSAIDs inhibits chaperone-mediated autophagy and induces hepatic lipid accumulation[J]. Theranostics, 2022, 12( 5): 2351- 2369. DOI: 10.7150/thno.70692. |
| [54] |
QIAO L, HU J, QIU X, et al. LAMP2A, LAMP2B and LAMP2C: similar structures, divergent roles[J]. Autophagy, 2023, 19( 11): 2837- 2852. DOI: 10.1080/15548627.2023.2235196. |
| [55] |
SCHULZE RJ, KRUEGER EW, WELLER SG, et al. Direct lysosome-based autophagy of lipid droplets in hepatocytes[J]. Proc Natl Acad Sci U S A, 2020, 117( 51): 32443- 32452. DOI: 10.1073/pnas.2011442117. |
| [56] |
LIAO PC, GARCIA EJ, TAN G, et al. Roles for L o microdomains and ESCRT in ER stress-induced lipid droplet microautophagy in budding yeast[J]. Mol Biol Cell, 2021, 32( 22): br12. DOI: 10.1091/mbc.E21-04-0179. |
| [57] |
GARCIA EJ, LIAO PC, TAN G, et al. Membrane dynamics and protein targets of lipid droplet microautophagy during ER stress-induced proteostasis in the budding yeast, Saccharomyces cerevisiae[J]. Autophagy, 2021, 17( 9): 2363- 2383. DOI: 10.1080/15548627.2020.1826691. |



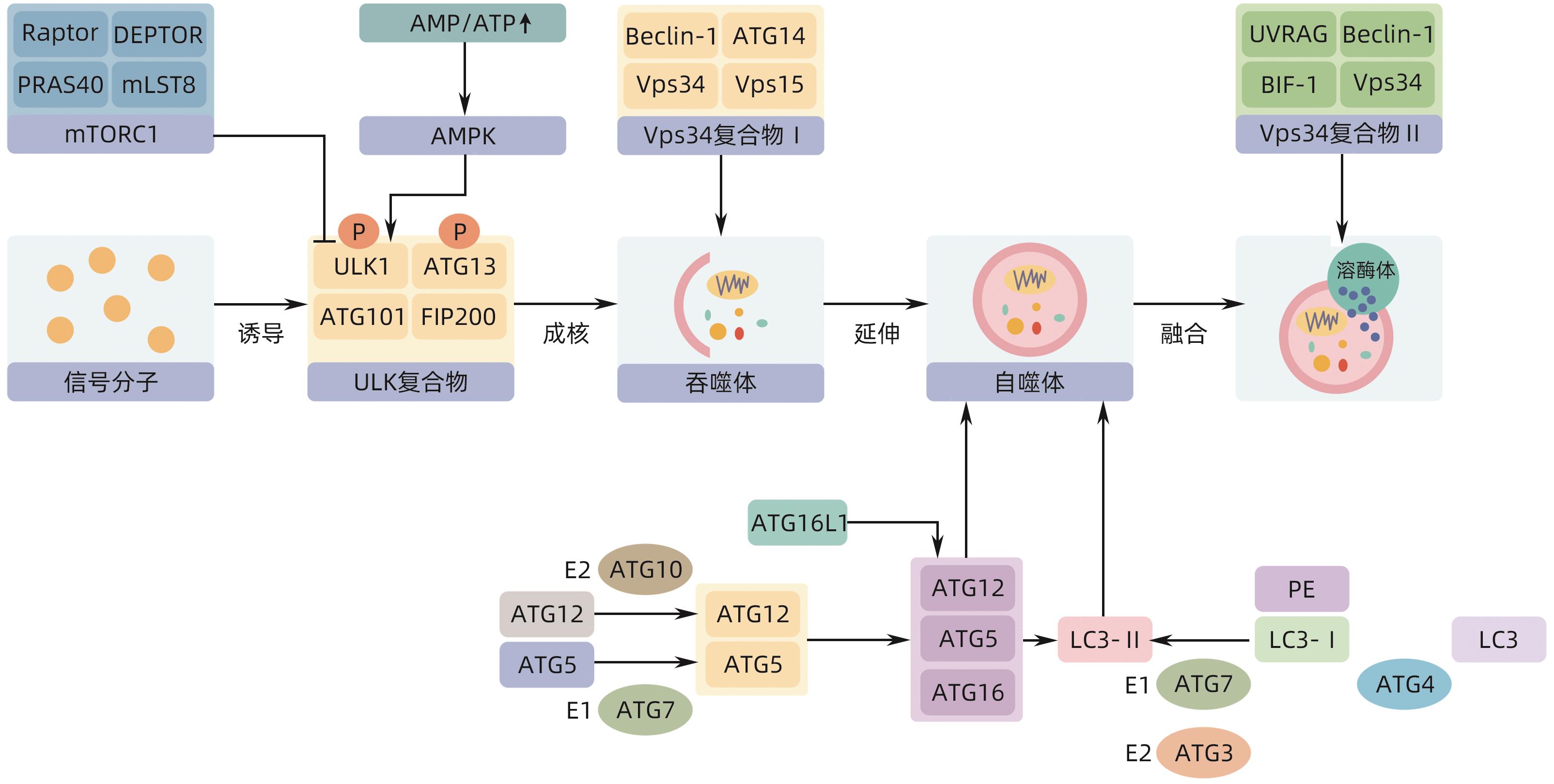




 DownLoad:
DownLoad:
