| [1] |
MORIS D, PALTA M, KIM C, et al. Advances in the treatment of intrahepatic cholangiocarcinoma: An overview of the current and future therapeutic landscape for clinicians[J]. CA Cancer J Clin, 2023, 73( 2): 198- 222. DOI: 10.3322/caac.21759. |
| [2] |
BERTUCCIO P, MALVEZZI M, CARIOLI G, et al. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma[J]. J Hepatol, 2019, 71( 1): 104- 114. DOI: 10.1016/j.jhep.2019.03.013. |
| [3] |
SUNG H, FERLAY J, SIEGEL R L, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71( 3): 209- 249. DOI: org/10.3322/caac.21660. |
| [4] |
RUMGAY H, ARNOLD M, FERLAY J, et al. Global burden of primary liver cancer in 2020 and predictions to 2040[J]. J Hepatol, 2022, 77( 6): 1598- 1606. DOI: 10.1016/j.jhep.2022.08.021. |
| [5] |
TAVOLARI S, BRANDI G. Mutational landscape of cholangiocarcinoma according to different etiologies: A review[J]. Cells, 2023, 12( 9): 1216. DOI: 10.3390/cells12091216. |
| [6] |
SONG ZM, LIN SR, WU XW, et al. Hepatitis B virus-related intrahepatic cholangiocarcinoma originates from hepatocytes[J]. Hepatol Int, 2023, 17( 5): 1300- 1317. DOI: 10.1007/s12072-023-10556-3. |
| [7] |
SLAGLE BL, BOUCHARD MJ. Role of HBx in hepatitis B virus persistence and its therapeutic implications[J]. Curr Opin Virol, 2018, 30: 32- 38. DOI: 10.1016/j.coviro.2018.01.007. |
| [8] |
RIVIÈRE L, GEROSSIER L, DUCROUX A, et al. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase[J]. J Hepatol, 2015, 63( 5): 1093- 1102. DOI: 10.1016/j.jhep.2015.06.023. |
| [9] |
DECORSIÈRE A, MUELLER H, van BREUGEL PC, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor[J]. Nature, 2016, 531( 7594): 386- 389. DOI: 10.1038/nature17170. |
| [10] |
BELLONI L, POLLICINO T, de NICOLA F, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function[J]. Proc Natl Acad Sci USA, 2009, 106( 47): 19975- 19979. DOI: 10.1073/pnas.0908365106. |
| [11] |
SONG GJ, YANG RF, JIN Q, et al. HBV pregenome RNA as a predictor of spontanous HBeAg seroconversion in HBeAg-positive chronic hepatitis B patients[J]. BMC Gastroenterol, 2023, 23( 1): 381. DOI: 10.1186/s12876-023-03023-8. |
| [12] |
LEE WY, BACHTIAR M, CHOO CCS, et al. Comprehensive review of Hepatitis B Virus-associated hepatocellular carcinoma research through text mining and big data analytics[J]. Biol Rev Camb Philos Soc, 2019, 94( 2): 353- 367. DOI: 10.1111/brv.12457. |
| [13] |
ZHOU YM, CAO L, LI B, et al. Expression of HBx protein in hepatitis B virus-infected intrahepatic cholangiocarcinoma[J]. Hepatobiliary Pancreat Dis Int, 2012, 11( 5): 532- 535. DOI: 10.1016/s1499-3872(12)60219-7. |
| [14] |
COUGOT D, ALLEMAND E, RIVIÈRE L, et al. Inhibition of PP1 phosphatase activity by HBx: A mechanism for the activation of hepatitis B virus transcription[J]. Sci Signal, 2012, 5( 205): ra1. DOI: 10.1126/scisignal.2001906. |
| [15] |
KIM WH, HONG F, JARUGA B, et al. Hepatitis B virus X protein sensitizes primary mouse hepatocytes to ethanol- and TNF-alpha-induced apoptosis by a caspase-3-dependent mechanism[J]. Cell Mol Immunol, 2005, 2( 1): 40- 48.
|
| [16] |
PAN JB, DUAN LX, SUN BS, et al. Hepatitis B virus X protein protects against anti-Fas-mediated apoptosis in human liver cells by inducing NF-kappa B[J]. J Gen Virol, 2001, 82( Pt 1): 171- 182. DOI: 10.1099/0022-1317-82-1-171. |
| [17] |
WANG P, GUO QS, WANG ZW, et al. HBx induces HepG-2 cells autophagy through PI3K/Akt-mTOR pathway[J]. Mol Cell Biochem, 2013, 372( 1-2): 161- 168. DOI: 10.1007/s11010-012-1457-x. |
| [18] |
WANG XY, WEI ZQ, CHENG B, et al. Endoplasmic reticulum stress promotes HBV production by enhancing use of the autophagosome/multivesicular body axis[J]. Hepatology, 2022, 75( 2): 438- 454. DOI: 10.1002/hep.32178. |
| [19] |
LIN Y, WU CC, WANG XY, et al. Hepatitis B virus is degraded by autophagosome-lysosome fusion mediated by Rab7 and related components[J]. Protein Cell, 2019, 10( 1): 60- 66. DOI: 10.1007/s13238-018-0555-2. |
| [20] |
ZHANG H, ZHANG YQ, ZHU XY, et al. DEAD box protein 5 inhibits liver tumorigenesis by stimulating autophagy via interaction with p62/SQSTM1[J]. Hepatology, 2019, 69( 3): 1046- 1063. DOI: 10.1002/hep.30300. |
| [21] |
YAN Y, LIU N, LU L, et al. Autophagy enhances antitumor immune responses induced by irradiated hepatocellular carcinoma cells engineered to express hepatitis B virus X protein[J]. Oncol Rep, 2013, 30( 2): 993- 999. DOI: 10.3892/or.2013.2531. |
| [22] |
SANZ-CAMENO P, MARTÍN-VÍLCHEZ S, LARA-PEZZI E, et al. Hepatitis B virus promotes angiopoietin-2 expression in liver tissue: Role of HBV x protein[J]. Am J Pathol, 2006, 169( 4): 1215- 1222. DOI: 10.2353/ajpath.2006.051246. |
| [23] |
LEVRERO M, ZUCMAN-ROSSI J. Mechanisms of HBV-induced hepatocellular carcinoma[J]. J Hepatol, 2016, 64( 1 Suppl): S84- S101. DOI: 10.1016/j.jhep.2016.02.021. |
| [24] |
ZOU SQ, QU ZL, LI ZF, et al. Hepatitis B virus X gene induces human telomerase reverse transcriptase mRNA expression in cultured normal human cholangiocytes[J]. World J Gastroenterol, 2004, 10( 15): 2259- 2262. DOI: 10.3748/wjg.v10.i15.2259. |
| [25] |
SALERNO D, CHIODO L, ALFANO V, et al. Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription[J]. Gut, 2020, 69( 11): 2016- 2024. DOI: 10.1136/gutjnl-2019-319637. |
| [26] |
DING WB, WANG MC, YU J, et al. HBV/pregenomic RNA increases the stemness and promotes the development of HBV-related HCC through reciprocal regulation with insulin-like growth factor 2 mRNA-binding protein 3[J]. Hepatology, 2021, 74( 3): 1480- 1495. DOI: 10.1002/hep.31850. |
| [27] |
ZHENG DL, ZHANG L, CHENG N, et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A[J]. J Hepatol, 2009, 50( 2): 377- 387. DOI: 10.1016/j.jhep.2008.10.019. |
| [28] |
CHEN YY, WANG WH, CHE L, et al. BNIP3L-dependent mitophagy promotes HBx-induced cancer stemness of hepatocellular carcinoma cells via glycolysis metabolism reprogramming[J]. Cancers, 2020, 12( 3): 655. DOI: 10.3390/cancers12030655. |
| [29] |
CHEN SL, ZHANG CZ, LIU LL, et al. A GYS2/p53 negative feedback loop restricts tumor growth in HBV-related hepatocellular carcinoma[J]. Cancer Res, 2019, 79( 3): 534- 545. DOI: 10.1158/0008-5472.CAN-18-2357. |
| [30] |
ZHAO LH, WANG Y, TIAN T, et al. Analysis of viral integration reveals new insights of oncogenic mechanism in HBV-infected intrahepatic cholangiocarcinoma and combined hepatocellular-cholangiocarcinoma[J]. Hepatol Int, 2022, 16( 6): 1339- 1352. DOI: 10.1007/s12072-022-10419-3. |
| [31] |
MASON WS, GILL US, LITWIN S, et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant[J]. Gastroenterology, 2016, 151( 5): 986- 998.e4. DOI: 10.1053/j.gastro.2016.07.012. |
| [32] |
AN J, KIM D, OH B, et al. Comprehensive characterization of viral integrations and genomic aberrations in HBV-infected intrahepatic cholangiocarcinomas[J]. Hepatology, 2022, 75( 4): 997- 1011. DOI.org/10.1002/hep.32135.
|
| [33] |
ZHAO LH, LIU X, YAN HX, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma[J]. Nat Commun, 2016, 7: 12992. DOI: 10.1038/ncomms12992. |
| [34] |
JANG JW, KIM HS, KIM JS, et al. Distinct patterns of HBV integration and TERT alterations between in tumor and non-tumor tissue in patients with hepatocellular carcinoma[J]. Int J Mol Sci, 2021, 22( 13): 7056. DOI: 10.3390/ijms22137056. |
| [35] |
LI XJ, ZHANG JB, YANG ZW, et al. The function of targeted host genes determines the oncogenicity of HBV integration in hepatocellular carcinoma[J]. J Hepatol, 2014, 60( 5): 975- 984. DOI: 10.1016/j.jhep.2013.12.014. |
| [36] |
HUANG XL, YU D, GU XT, et al. A comparative study of clinicopathological and imaging features of HBV-negative and HBV-positive intrahepatic cholangiocarcinoma patients with different pathologic differentiation degrees[J]. Sci Rep, 2023, 13( 1): 19726. DOI: 10.1038/s41598-023-47108-6. |
| [37] |
ZOU SS, LI JR, ZHOU HB, et al. Mutational landscape of intrahepatic cholangiocarcinoma[J]. Nat Commun, 2014, 5: 5696. DOI: 10.1038/ncomms6696. |
| [38] |
SZE KMF, CHU GKY, LEE JMF, et al. C-terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma[J]. Hepatology, 2013, 57( 1): 131- 139. DOI: 10.1002/hep.25979. |
| [39] |
FRAGKOU N, SIDERAS L, PANAS P, et al. Update on the association of hepatitis B with intrahepatic cholangiocarcinoma: Is there new evidence?[J]. World J Gastroenterol, 2021, 27( 27): 4252- 4275. DOI: 10.3748/wjg.v27.i27.4252. |
| [40] |
GATSELIS NK, TEPETES K, LOUKOPOULOS A, et al. Hepatitis B virus and intrahepatic cholangiocarcinoma[J]. Cancer Investig, 2007, 25( 1): 55- 58. DOI: 10.1080/07357900601130722. |
| [41] |
CALADO RT, BRUDNO J, MEHTA P, et al. Constitutional telomerase mutations are genetic risk factors for cirrhosis[J]. Hepatology, 2011, 53( 5): 1600- 1607. DOI: 10.1002/hep.24173. |
| [42] |
SHEN YF, XU SS, YE CQ, et al. Proteomic and single-cell landscape reveals novel pathogenic mechanisms of HBV-infected intrahepatic cholangiocarcinoma[J]. iScience, 2023, 26( 2): 106003. DOI: 10.1016/j.isci.2023.106003. |
| [43] |
ILYAS SI, AFFO S, GOYAL L, et al. Cholangiocarcinoma-novel biological insights and therapeutic strategies[J]. Nat Rev Clin Oncol, 2023, 20( 7): 470- 486. DOI: 10.1038/s41571-023-00770-1. |
| [44] |
GONG J, LIANG YL, ZHOU WY, et al. Prognostic value of neutrophil-to-lymphocyte ratio associated with prognosis in HBV-infected patients[J]. J Med Virol, 2018, 90( 4): 730- 735. DOI: 10.1002/jmv.25015. |
| [45] |
ZHENG BH, MA JQ, TIAN LY, et al. The distribution of immune cells within combined hepatocellular carcinoma and cholangiocarcinoma predicts clinical outcome[J]. Clin Transl Med, 2020, 10( 1): 45- 56. DOI: 10.1002/ctm2.11. |
| [46] |
MA BQ, MENG HJ, TIAN Y, et al. Distinct clinical and prognostic implication of IDH1/2 mutation and other most frequent mutations in large duct and small duct subtypes of intrahepatic cholangiocarcinoma[J]. BMC Cancer, 2020, 20( 1): 318. DOI: 10.1186/s12885-020-06804-6. |
| [47] |
KOHANBASH G, CARRERA DA, SHRIVASTAV S, et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8 + T cell accumulation in gliomas[J]. J Clin Invest, 2017, 127( 4): 1425- 1437. DOI: 10.1172/JCI90644. |
| [48] |
KENDALL T, VERHEIJ J, GAUDIO E, et al. Anatomical, histomorphological and molecular classification of cholangiocarcinoma[J]. Liver Int, 2019, 39( Suppl 1): 7- 18. DOI: 10.1111/liv.14093. |
| [49] |
TJWA ETTL, VAN OORD GW, HEGMANS JP, et al. Viral load reduction improves activation and function of natural killer cells in patients with chronic hepatitis B[J]. J Hepatol, 2011, 54( 2): 209- 218. DOI: 10.1016/j.jhep.2010.07.009. |
| [50] |
FISICARO P, BARILI V, MONTANINI B, et al. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B[J]. Nat Med, 2017, 23( 3): 327- 336. DOI: 10.1038/nm.4275. |
| [51] |
TENG CF, LI TC, WANG T, et al. Increased infiltration of regulatory T cells in hepatocellular carcinoma of patients with hepatitis B virus pre-S2 mutant[J]. Sci Rep, 2021, 11( 1): 1136. DOI: 10.1038/s41598-020-80935-5. |
| [52] |
YU X, LAN PX, HOU XB, et al. HBV inhibits LPS-induced NLRP3 inflammasome activation and IL-1β production via suppressing the NF-κB pathway and ROS production[J]. J Hepatol, 2017, 66( 4): 693- 702. DOI: 10.1016/j.jhep.2016.12.018. |
| [53] |
LI M, ZHOU ZH, SUN XH, et al. Hepatitis B core antigen upregulates B7-H1 on dendritic cells by activating the AKT/ERK/P38 pathway: A possible mechanism of hepatitis B virus persistence[J]. Lab Invest, 2016, 96( 11): 1156- 1164. DOI: 10.1038/labinvest.2016.96. |
| [54] |
ZHOU L, HE R, FANG PN, et al. Hepatitis B virus rigs the cellular metabolome to avoid innate immune recognition[J]. Nat Commun, 2021, 12( 1): 98. DOI: 10.1038/s41467-020-20316-8. |
| [55] |
FISICARO P, BONI C, BARILI V, et al. Strategies to overcome HBV-specific T cell exhaustion: Checkpoint inhibitors and metabolic re-programming[J]. Curr Opin Virol, 2018, 30: 1- 8. DOI: 10.1016/j.coviro.2018.01.003. |
| [56] |
YAN FN, ZHANG Q, SHI K, et al. Gut microbiota dysbiosis with hepatitis B virus liver disease and association with immune response[J]. Front Cell Infect Microbiol, 2023, 13: 1152987. DOI: 10.3389/fcimb.2023.1152987. |



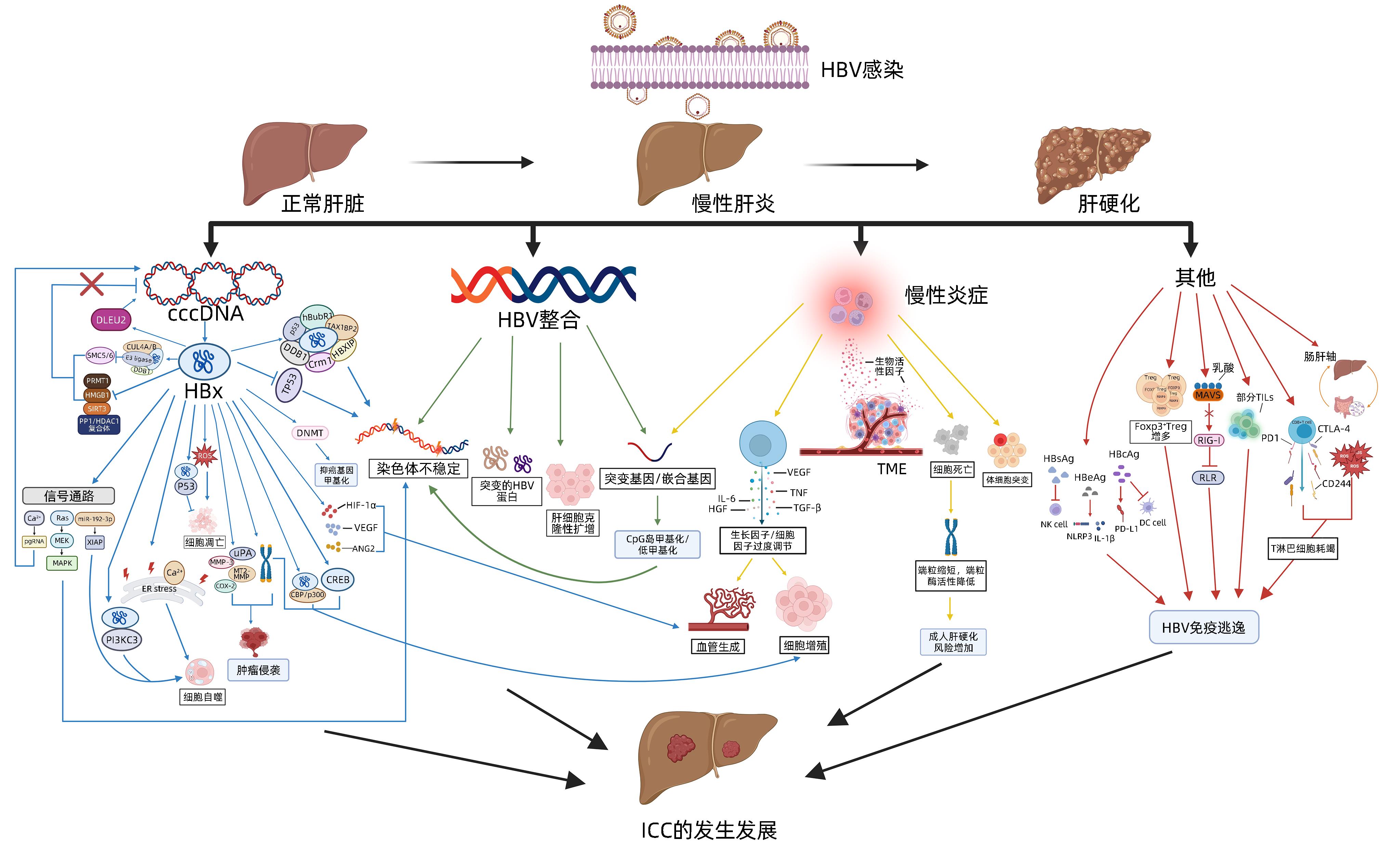




 DownLoad:
DownLoad: