乙型肝炎动物模型的研究现状与挑战
DOI: 10.12449/JCH260105
-
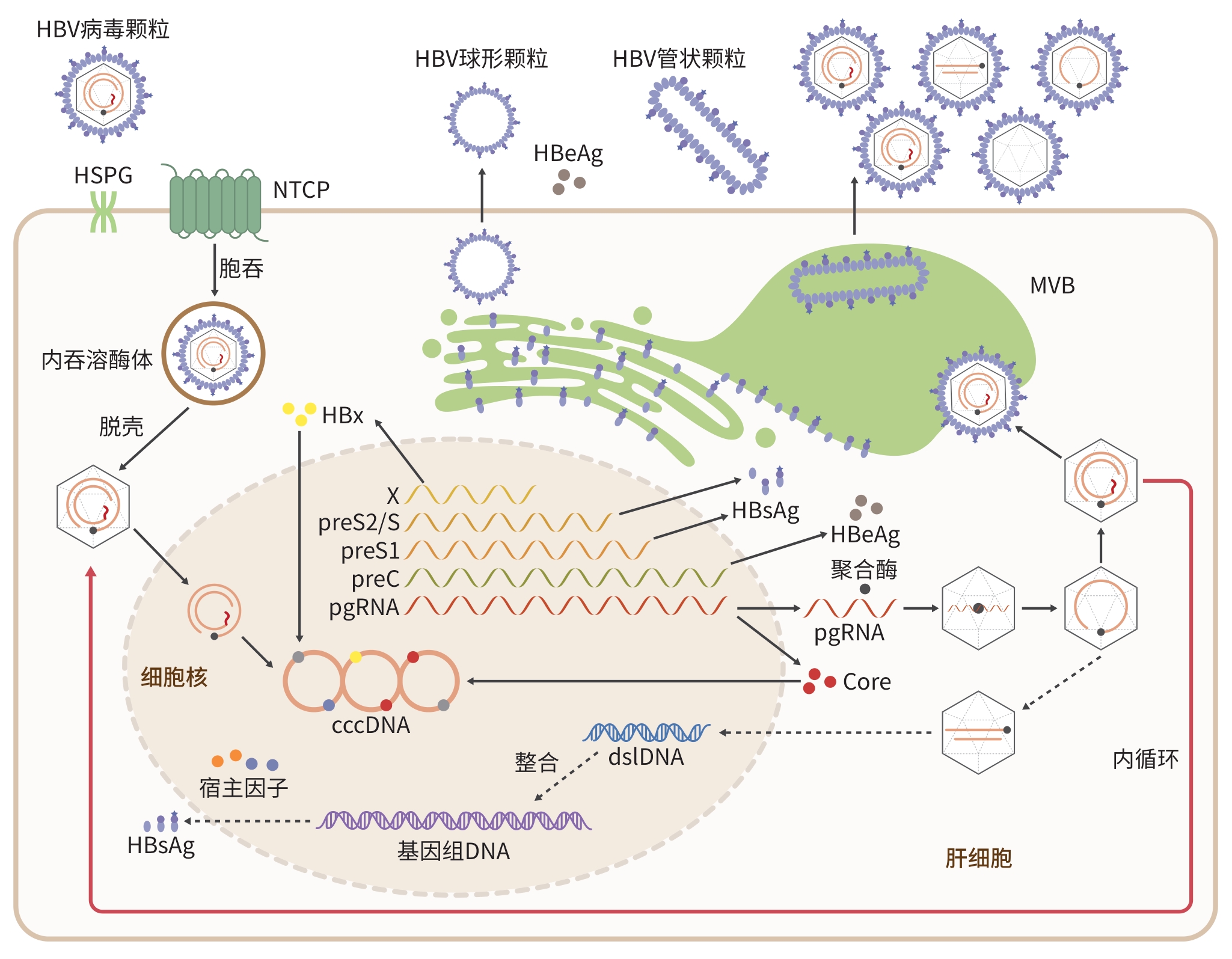
摘要: 乙型肝炎病毒(HBV)感染是全球面临的主要健康问题之一,是导致肝硬化和肝细胞癌的重要病因之一。由于HBV感染具有严格的种属特异性,目前尚未建立能够支持完整HBV感染周期并真实模拟宿主免疫和发病机制的动物模型。现有用于HBV研究的动物模型包括黑猩猩、树鼩和小鼠等多种宿主,以及利用相关嗜肝病毒建立的替代模型。尽管这些模型在研究HBV复制、免疫反应及抗病毒药物评估方面发挥了重要作用,但仍存在伦理、感染效率低、成本高或缺乏持续感染等局限。近年来,人源化肝脏与免疫系统的小鼠模型、转基因模型及病毒载体介导的感染模型等新策略的出现,显著推动了HBV生物学研究进展。未来,通过结合基因编辑、组织工程及多系统整合等新技术,构建更符合人类病理生理特征的HBV感染动物模型,将为深入理解病毒-宿主互作机制、探索HBV清除途径及开发根治性治疗策略提供坚实基础。Abstract: Hepatitis B virus (HBV) infection is one of the major global health problems, and it can lead to the development of liver cirrhosis and hepatocellular carcinoma. Due to the strict species specificity of HBV infection, no animal model has yet been established to fully support the complete life cycle of HBV infection and accurately reflect host immune responses and pathogenesis. Current animal models used for HBV research include various hosts such as chimpanzees, tree shrews, and mice, as well as surrogate models based on related hepatotropic viruses. Although these models have contributed significantly to the research on HBV replication, immune response, and antiviral drug evaluation, they still have certain limitations such as ethical concerns, low infection efficiency, high costs, and a lack of persistent infection. In recent years, the development of novel strategies, such as humanized mouse models with reconstituted human liver and immune systems, transgenic models, and viral vector-mediated infection systems, has greatly promoted the research on HBV biology. In the future, with the integration of emerging technologies including gene editing, tissue engineering, and multi-system reconstruction, it is possible to establish HBV infection models that can more closely mimic human pathophysiology, thereby laying a robust foundation for understanding virus-host interactions, exploring the pathways for viral clearance, and developing radical treatment strategies.
-
Key words:
- Hepatitis B Virus /
- Covalently Closed Circular DNA /
- Models, Animal
-
表 1 不同物种肝细胞对HBV感染与复制的支持能力
Table 1. Ability of hepatocytes from different species to support HBV infection and replication
物种 HBV进入 cccDNA形成及病毒
复制参考文献 人 + + [3,11-13] 黑猩猩 + + [14-17] 树鼩 + + [3,13] 土拨鼠 + - [13] 猕猴 - + [3,13,18] 食蟹猴 - + [3,11-12] 马 + + [13] 牛 +/- - [12-13] 山羊 +/- - [12-13] 猫 + + [12-13] 兔 + - [12-13,19] 仓鼠 - + [12-13,20] 豚鼠 - - [12] 猪 - + [11-12,19] 狗 + - [11-12] 大鼠 + - [11] 小鼠 - - [10-11,21] 注:“+”表示有;“-”表示无;“+/-”表示现有数据存在冲突。HBV,乙型肝炎病毒;cccDNA,共价闭合环状DNA。
表 2 HBV小鼠模型
Table 2. HBV mice model
小鼠模型 载体/病毒
类型病毒
进入感染性 病毒
复制病毒持
久性cccDNA 病毒颗
粒分泌免疫系统 优势 局限性 HBV转基因
小鼠整合HBV
基因- - + + - + 免疫完全 易获取;近交
系;稳定表达
HBV基因非自然感染;无
cccDNA形成;无
病毒清除;免疫
耐受高压水动力
注射介导的
HBV转染小鼠HBV复制
型质粒- - + + - + 免疫完全 易获取;近交
系;研究病毒
清除机制非自然感染;瞬
时载体驱动的复
制;转染效率低;
复制时间短重组AdV介
导的HBV
转导小鼠AdV-HBV - - + +/剂量依赖 - + 免疫完全 易获取;近交
系;稳定复制非自然感染;无
cccDNA形成;剂
量依赖重组AVV介
导的HBV转
导小鼠AAV-HBV - - + +/剂量和品
系依赖+ + 免疫完全 易获取;近交
系;稳定复制;
支持cccDNA
形成非自然感染;无
病毒清除;剂量
和品系依赖;免
疫耐受cccDNA
替代小鼠重组
cccDNA- - + + + + 免疫完全 易获取;近交
系;稳定复制;
以cccDNA或
其替代分子为
复制模板非自然感染 人-鼠嵌合肝
脏小鼠HBV + + + + + + 免疫缺陷 易获取;近交
系;HBV易感感染过程缓慢;
造模操作复杂;
繁殖困难;免疫
缺陷人免疫系统
和肝双人源
化小鼠HBV + + + + + + 人的免疫
系统易获取;近交
系;HBV易感造模操作复杂;
繁殖困难;成本
较高注:“+”表示有或包含;“-”表示无或没有。HBV,乙型肝炎病毒;cccDNA,共价闭合环状DNA;AdV,腺病毒;AAV,腺相关病毒。
-
[1] SCHWEITZER A, HORN J, MIKOLAJCZYK RT, et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: A systematic review of data published between 1965 and 2013[J]. Lancet, 2015, 386( 10003): 1546- 1555. DOI: 10.1016/S0140-6736(15)61412-X. [2] NASSAL M. HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B[J]. Gut, 2015, 64( 12): 1972- 1984. DOI: 10.1136/gutjnl-2015-309809. [3] YAN H, ZHONG GC, XU GW, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus[J]. eLife, 2012, 1: e00049. DOI: 10.7554/eLife.00049. [4] XIA YC, GUO HT. Hepatitis B virus cccDNA: Formation, regulation and therapeutic potential[J]. Antiviral Res, 2020, 180: 104824. DOI: 10.1016/j.antiviral.2020.104824. [5] ZHAO KT, WANG JJ, WANG ZC, et al. Hepatitis B virus hijacks MRE11-RAD50-NBS1 complex to form its minichromosome[J]. PLoS Pathog, 2025, 21( 1): e1012824. DOI: 10.1371/journal.ppat.1012824. [6] XIA YC, CHENG XM, NILSSON T, et al. Nucleolin binds to and regulates transcription of hepatitis B virus covalently closed circular DNA minichromosome[J]. Proc Natl Acad Sci USA, 2023, 120( 49): e2306390120. DOI: 10.1073/pnas.2306390120. [7] TENG Y, XU ZC, ZHAO KT, et al. Novel function of SART1 in HNF4α transcriptional regulation contributes to its antiviral role during HBV infection[J]. J Hepatol, 2021, 75( 5): 1072- 1082. DOI: 10.1016/j.jhep.2021.06.038. [8] WANG JJ, HUANG HY, ZHAO KT, et al. G-quadruplex in hepatitis B virus pregenomic RNA promotes its translation[J]. J Biol Chem, 2023, 299( 9): 105151. DOI: 10.1016/j.jbc.2023.105151. [9] ZHENG YC, WANG MF, LI ST, et al. Hepatitis B virus hijacks TSG101 to facilitate egress via multiple vesicle bodies[J]. PLoS Pathog, 2023, 19( 5): e1011382. DOI: 10.1371/journal.ppat.1011382. [10] ZHAO KT, GUO FT, WANG JJ, et al. Limited disassembly of cytoplasmic hepatitis B virus nucleocapsids restricts viral infection in murine hepatic cells[J]. Hepatology, 2023, 77( 4): 1366- 1381. DOI: 10.1002/hep.32622. [11] LEMPP FA, WIEDTKE E, QU BQ, et al. Sodium taurocholate cotransporting polypeptide is the limiting host factor of hepatitis B virus infection in macaque and pig hepatocytes[J]. Hepatology, 2017, 66( 3): 703- 716. DOI: 10.1002/hep.29112. [12] XU ZC, ZHAO KT, WANG JJ, et al. Screening of different species reveals cat hepatocytes support HBV infection[J]. PLoS Pathog, 2025, 21( 8): e1013390. DOI: 10.1371/journal.ppat.1013390. [13] CHEN FW, WETTENGEL JM, GEGENFURTNER F, et al. Identification of NTCP animal orthologs supporting hepatitis B virus binding and infection[J]. J Virol, 2025, 99( 4): e01833-24. DOI: 10.1128/jvi.01833-24. [14] ASABE S, WIELAND SF, CHATTOPADHYAY PK, et al. The size of the viral inoculum contributes to the outcome of hepatitis B virus infection[J]. J Virol, 2009, 83( 19): 9652- 9662. DOI: 10.1128/JVI.00867-09. [15] WIELAND SF, SPANGENBERG HC, THIMME R, et al. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees[J]. Proc Natl Acad Sci USA, 2004, 101( 7): 2129- 2134. DOI: 10.1073/pnas.0308478100. [16] GUIDOTTI LG, ROCHFORD R, CHUNG J, et al. Viral clearance without destruction of infected cells during acute HBV infection[J]. Science, 1999, 284( 5415): 825- 829. DOI: 10.1126/science.284.5415.825. [17] KOMIYA Y, KATAYAMA K, YUGI H, et al. Minimum infectious dose of hepatitis B virus in chimpanzees and difference in the dynamics of viremia between genotype A and genotype C[J]. Transfusion, 2008, 48( 2): 286- 294. DOI: 10.1111/j.1537-2995.2007.01522.x. [18] BURWITZ BJ, WETTENGEL JM, MÜCK-HÄUSL MA, et al. Hepatocytic expression of human sodium-taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques[J]. Nat Commun, 2017, 8( 1): 2146. DOI: 10.1038/s41467-017-01953-y. [19] ZHOU M, QIN B, DENG XS, et al. hNTCP-expressing primary pig hepatocytes are a valuable tool for investigating hepatitis B virus infection and antiviral drugs[J]. Mol Med Rep, 2019, 20( 4): 3820- 3828. DOI: 10.3892/mmr.2019.10628. [20] ZHANG H, LIU YN, LIU CD, et al. The feasibility of establishing a Hamster model for HBV infection: In vitro evidence[J]. mBio, 2024, 15( 11): e02615-24. DOI: 10.1128/mbio.02615-24. [21] YAN H, PENG B, HE WH, et al. Molecular determinants of hepatitis B and D virus entry restriction in mouse sodium taurocholate cotransporting polypeptide[J]. J Virol, 2013, 87( 14): 7977- 7991. DOI: 10.1128/JVI.03540-12. [22] WANG Q, SCHWARZENBERGER P, YANG F, et al. Experimental chronic hepatitis B infection of neonatal tree shrews(Tupaia belangeri Chinensis): A model to study molecular causes for susceptibility and disease progression to chronic hepatitis in humans[J]. Virol J, 2012, 9: 170. DOI: 10.1186/1743-422X-9-170. [23] KÖCK J, NASSAL M, MACNELLY S, et al. Efficient infection of primary Tupaia Hepatocytes with purified human and woolly monkey hepatitis B virus[J]. J Virol, 2001, 75( 11): 5084- 5089. DOI: 10.1128/JVI.75.11.5084-5089.2001. [24] GHEIT T, SEKKAT S, COVA L, et al. Experimental transfection of Macaca sylvanus with cloned human hepatitis B virus[J]. J Gen Virol, 2002, 83( Pt 7): 1645- 1649. DOI: 10.1099/0022-1317-83-7-1645. [25] MÜLLER SF, KÖNIG A, DÖRING B, et al. Characterisation of the hepatitis B virus cross-species transmission pattern via Na+/taurocholate co-transporting polypeptides from 11 New World and Old World primate species[J]. PLoS One, 2018, 13( 6): e0199200. DOI: 10.1371/journal.pone.0199200. [26] BISWAS S, RUST LN, WETTENGEL JM, et al. Long-term hepatitis B virus infection of Rhesus macaques requires suppression of host immunity[J]. Nat Commun, 2022, 13( 1): 2995. DOI: 10.1038/s41467-022-30593-0. [27] RUST LN, WETTENGEL JM, BISWAS S, et al. Liver-specific transgenic expression of human NTCP in Rhesus macaques confers HBV susceptibility on primary hepatocytes[J]. Proc Natl Acad Sci USA, 2025, 122( 7): e2413771122. DOI: 10.1073/pnas.2413771122. [28] HE WH, REN BJ, MAO FF, et al. Hepatitis D virus infection of mice expressing human sodium taurocholate co-transporting polypeptide[J]. PLoS Pathog, 2015, 11( 4): e1004840. DOI: 10.1371/journal.ppat.1004840. [29] GUIDOTTI LG, MATZKE B, SCHALLER H, et al. High-level hepatitis B virus replication in transgenic mice[J]. J Virol, 1995, 69( 10): 6158- 6169. DOI: 10.1128/JVI.69.10.6158-6169.1995. [30] YANG PL, ALTHAGE A, CHUNG J, et al. Hydrodynamic injection of viral DNA: A mouse model of acute hepatitis B virus infection[J]. Proc Natl Acad Sci USA, 2002, 99( 21): 13825- 13830. DOI: 10.1073/pnas.202398599. [31] SPRENGERS D, VAN DER MOLEN RG, KUSTERS JG, et al. Analysis of intrahepatic HBV-specific cytotoxic T-cells during and after acute HBV infection in humans[J]. J Hepatol, 2006, 45( 2): 182- 189. DOI: 10.1016/j.jhep.2005.12.022. [32] YANG D, LIU LC, ZHU DM, et al. A mouse model for HBV immunotolerance and immunotherapy[J]. Cell Mol Immunol, 2014, 11( 1): 71- 78. DOI: 10.1038/cmi.2013.43. [33] KO C, SU JP, FESTAG J, et al. Intramolecular recombination enables the formation of hepatitis B virus(HBV) cccDNA in mice after HBV genome transfer using recombinant AAV vectors[J]. Antiviral Res, 2021, 194: 105140. DOI: 10.1016/j.antiviral.2021.105140. [34] QI ZH, LI GY, HU H, et al. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice[J]. J Virol, 2014, 88( 14): 8045- 8056. DOI: 10.1128/JVI.01024-14. [35] MA KAY, HE CY, CHEN ZY. A robust system for production of minicircle DNA vectors[J]. Nat Biotechnol, 2010, 28( 12): 1287- 1289. DOI: 10.1038/nbt.1708. [36] ZHOU ZM, LI C, TAN ZX, et al. A spatiotemporally controlled recombinant cccDNA mouse model for studying HBV and developing drugs against the virus[J]. Antiviral Res, 2023, 216: 105642. DOI: 10.1016/j.antiviral.2023.105642. [37] XU ZC, ZHAO L, ZHONG YQ, et al. A novel mouse model harboring hepatitis B virus covalently closed circular DNA[J]. Cell Mol Gastroenterol Hepatol, 2022, 13( 4): 1001- 1017. DOI: 10.1016/j.jcmgh.2021.11.011. [38] BILITY MT, ZHANG LG, WASHBURN ML, et al. Generation of a humanized mouse model with both human immune system and liver cells to model hepatitis C virus infection and liver immunopathogenesis[J]. Nat Protoc, 2012, 7( 9): 1608- 1617. DOI: 10.1038/nprot.2012.083. [39] WASHBURN ML, BILITY MT, ZHANG LG, et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease[J]. Gastroenterology, 2011, 140( 4): 1334- 1344. DOI: 10.1053/j.gastro.2011.01.001. [40] JESKE SD, WETTENGEL JM, GEGENFURTNER F, et al. Identification of amino acids restricting HBV receptor function in porcine NTCP[J]. NPJ Viruses, 2024, 2: 30. DOI: 10.1038/s44298-024-00041-5. [41] DUPINAY T, GHEIT T, ROQUES P, et al. Discovery of naturally occurring transmissible chronic hepatitis B virus infection among Macaca fascicularis from Mauritius Island[J]. Hepatology, 2013, 58( 5): 1610- 1620. DOI: 10.1002/hep.26428. [42] LANFORD RE, CHAVEZ D, BRASKY KM, et al. Isolation of a hepadnavirus from the woolly monkey, a New World primate[J]. Proc Natl Acad Sci USA, 1998, 95( 10): 5757- 5761. DOI: 10.1073/pnas.95.10.5757. [43] CHEN CY, WINER BY, CHAVEZ D, et al. Woolly monkey-HBV infection in squirrel monkeys as a surrogate nonhuman primate model of HBV infection[J]. Hepatol Commun, 2020, 4( 3): 371- 386. DOI: 10.1002/hep4.1471. [44] LIU YZ, CAFIERO TR, PARK D, et al. Targeted viral adaptation generates a Simian-tropic hepatitis B virus that infects marmoset cells[J]. Nat Commun, 2023, 14: 3582. DOI: 10.1038/s41467-023-39148-3. [45] de CARVALHO DOMINGUEZ SOUZA BF, KÖNIG A, RASCHE A, et al. A novel hepatitis B virus species discovered in capuchin monkeys sheds new light on the evolution of primate hepadnaviruses[J]. J Hepatol, 2018, 68( 6): 1114- 1122. DOI: 10.1016/j.jhep.2018.01.029. [46] SUMMERS J, SMOLEC JM, SNYDER R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks[J]. Proc Natl Acad Sci USA, 1978, 75( 9): 4533- 4537. DOI: 10.1073/pnas.75.9.4533. [47] DANDRI M, BURDA MR, WILL H, et al. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA[J]. Hepatology, 2000, 32( 1): 139- 146. DOI: 10.1053/jhep.2000.8701. [48] MENNE S, TUMAS DB, LIU KH, et al. Sustained efficacy and seroconversion with the Toll-like receptor 7 agonist GS-9620 in the Woodchuck model of chronic hepatitis B[J]. J Hepatol, 2015, 62( 6): 1237- 1245. DOI: 10.1016/j.jhep.2014.12.026. [49] MASON WS, SEAL G, SUMMERS J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus[J]. J Virol, 1980, 36( 3): 829- 836. DOI: 10.1128/JVI.36.3.829-836.1980. [50] GUO WN, ZHU B, AI L, et al. Animal models for the study of hepatitis B virus infection[J]. Zool Res, 2018, 39( 1): 25- 31. DOI: 10.24272/j.issn.2095-8137.2018.013. [51] FUNK A, MHAMDI M, WILL H, et al. Avian hepatitis B viruses: Molecular and cellular biology, phylogenesis, and host tropism[J]. World J Gastroenterol, 2007, 13( 1): 91- 103. DOI: 10.3748/wjg.v13.i1.91. [52] AGHAZADEH M, SHI M, BARRS VR, et al. A novel hepadnavirus identified in an immunocompromised domestic cat in Australia[J]. Viruses, 2018, 10( 5): 269. DOI: 10.3390/v10050269. [53] LANAVE G, CAPOZZA P, DIAKOUDI G, et al. Identification of hepadnavirus in the sera of cats[J]. Sci Rep, 2019, 9( 1): 10668. DOI: 10.1038/s41598-019-47175-8. [54] SHOFA M, OHKAWA A, KANEKO Y, et al. Conserved use of the sodium/bile acid cotransporter(NTCP) as an entry receptor by hepatitis B virus and domestic cat hepadnavirus[J]. Antiviral Res, 2023, 217: 105695. DOI: 10.1016/j.antiviral.2023.105695. [55] SHOFA M, FUKUSHIMA YV, SAITO A. Conserved yet divergent Smc5/6 complex degradation by mammalian hepatitis B virus X proteins[J]. Int J Mol Sci, 2025, 26( 14): 6786. DOI: 10.3390/ijms26146786. [56] CAVASIN JP, CHEN MC, AJOYAN H, et al. Recurrent integration of domestic cat hepatitis B virus DNA near feline CCNE1 supports an oncogenic role in hepatocellular carcinoma in cats[J]. Tumour Virus Res, 2025, 20: 200324. DOI: 10.1016/j.tvr.2025.200324. -



 PDF下载 ( 805 KB)
PDF下载 ( 805 KB)

 下载:
下载:


