HBV DNA聚合酶在介导HBV相关肝细胞癌肿瘤细胞免疫逃逸中的作用
DOI: 10.3969/j.issn.1001-5256.2023.12.017
Role of HBV DNA polymerase in mediating the immune escape of tumor cells in HBV-related hepatocellular carcinoma
-
摘要:
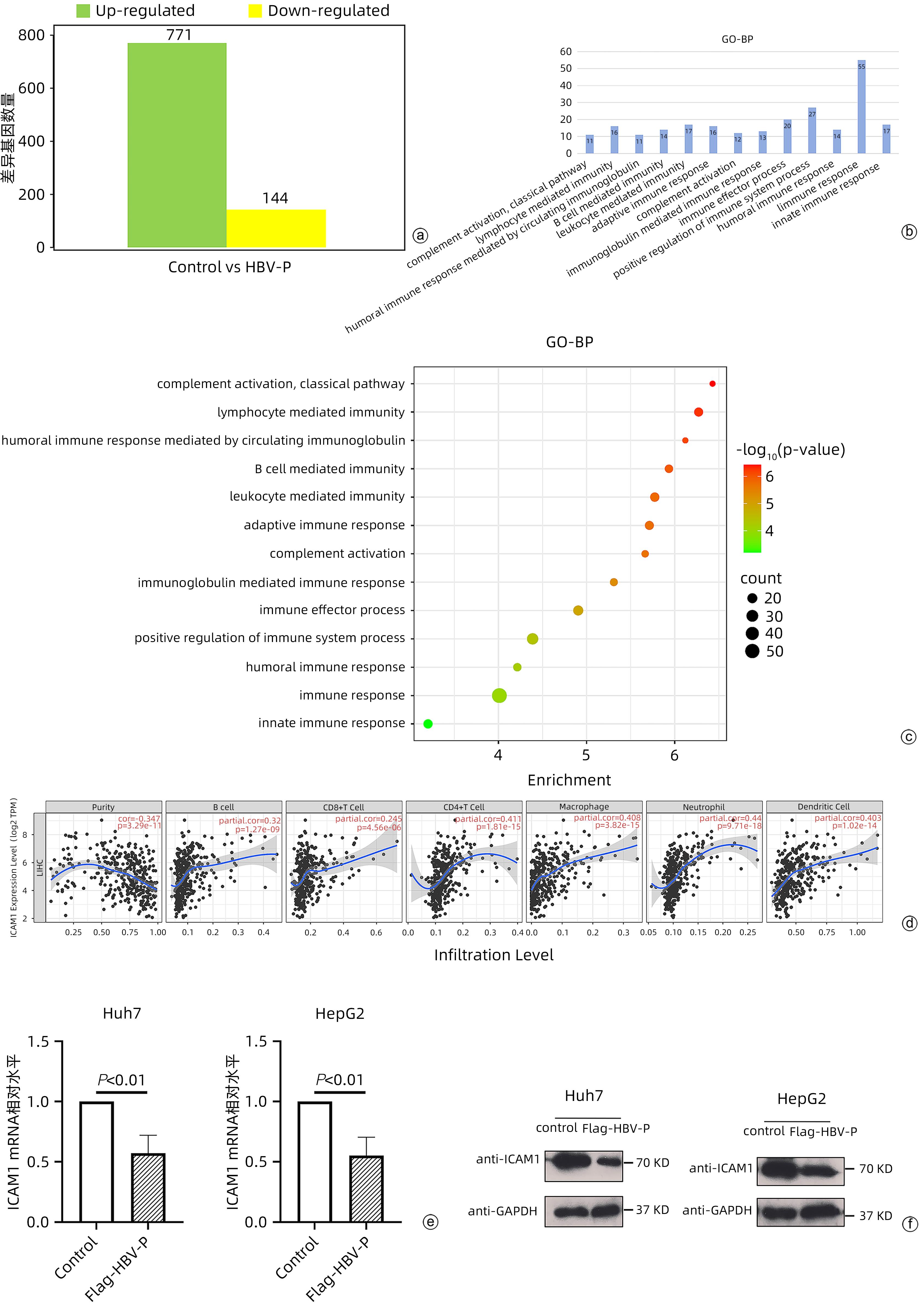
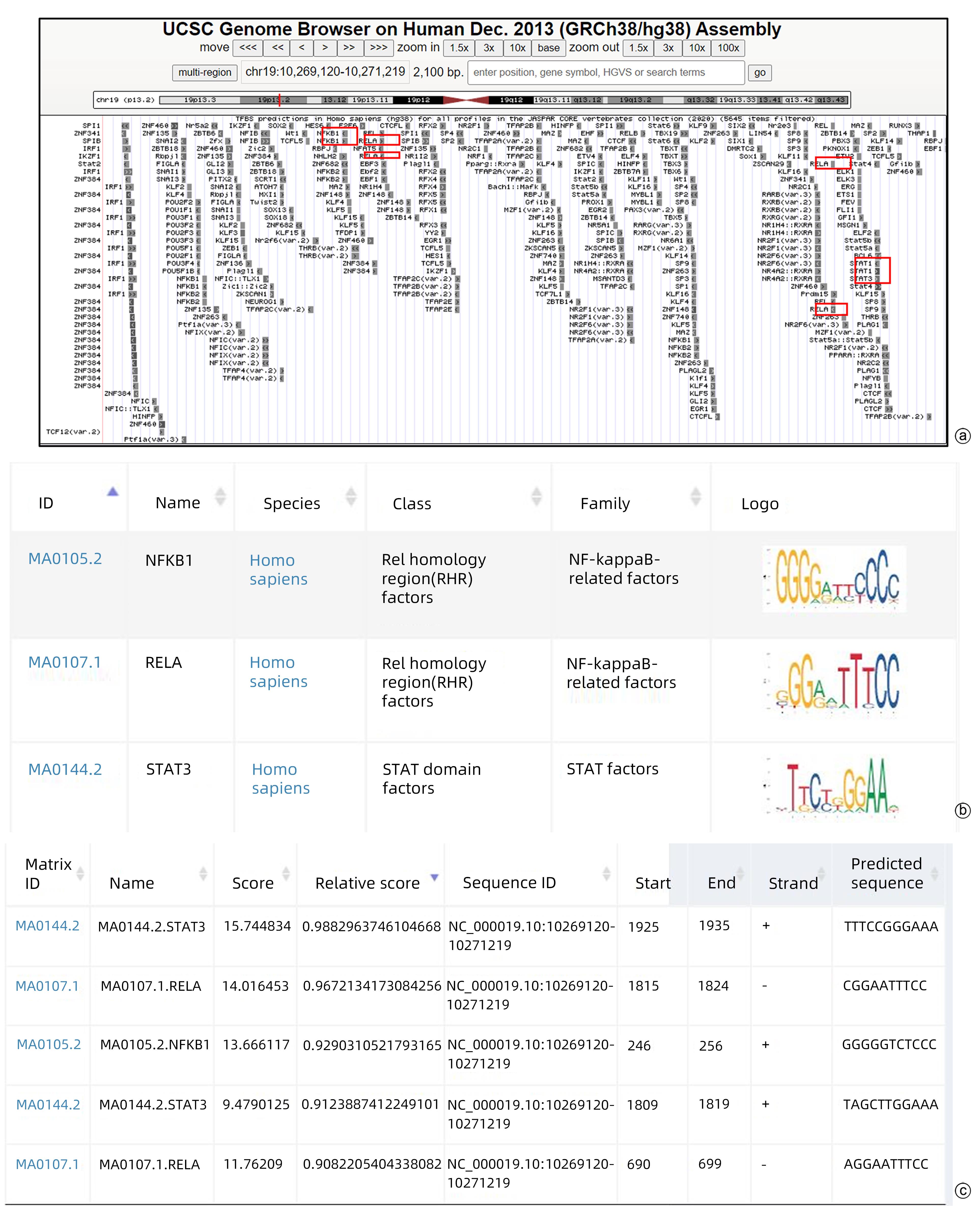
目的 本研究旨在明确HBV DNA聚合酶是否与T淋巴细胞功能衰竭相关,进而介导HBV相关肝细胞癌(HCC)肿瘤细胞的免疫逃逸以及探寻具体的分子机制。 方法 稳定转染带有Flag标签的HBV DNA聚合酶表达质粒(Flag-HBV-P)和细胞间黏附分子-1(ICAM1)的肝癌细胞系Huh7、HepG2与Jurkat细胞共培养,MTT、qRT-PCR、酶联免疫吸附测定(ELISA)实验分别检测Jurkat细胞增殖、活化(CD69表达)以及细胞因子IFN-γ分泌情况。RNA-seq筛选稳转细胞系与对照细胞中表达差异的免疫相关分子,mRNA半衰期及蛋白半衰期实验确定HBV DNA聚合酶对免疫相关分子具体在何种水平产生影响。网站预测此免疫相关分子启动子区域可能结合的转录因子,Western Blot实验验证转录因子对免疫相关分子的影响,挽救实验确定HBV DNA聚合酶是否通过此转录因子影响免疫相关分子的表达水平。两组间比较采用成组t检验。 结果 与对照组相比,实验组细胞(Huh7、HepG2)增殖、活化及细胞因子分泌明显降低(P值均<0.01)。与对照细胞相比,实验组细胞(Huh7、HepG2)的ICAM1 mRNA和蛋白水平均降低(P值均<0.01)。网站预测ICAM1启动子并初步筛选锚定了NFKB1、RELA和STAT3。与对照组相比,实验组细胞(Huh7、HepG2)p65蛋白的表达水平明显降低(P值均<0.01)。过表达p65后,ICAM1蛋白表达水平明显升高,降表达p65后,ICAM1蛋白表达水平明显降低(P值均<0.01),挽救实验中过表达p65后,对照组与实验组细胞的ICAM1表达水平无明显差异(P值均>0.05)。过表达ICAM1后,对照组与实验组细胞(Huh7、HepG2)的增殖、活化及细胞因子分泌无明显差异(P值均>0.05)。 结论 HBV DNA聚合酶通过抑制NF-κB中p65的表达来下调ICAM1的水平以介导HCC免疫逃逸。 -
关键词:
- 癌, 肝细胞 /
- 肿瘤逃逸 /
- 指导DNA的DNA聚合酶 /
- 细胞黏附分子
Abstract:Objective To determine whether HBV DNA polymerase is associated with T-cell failure and thus mediates the immune escape of HBV-related hepatocellular carcinoma (HCC) tumor cells, and to investigate the specific molecular mechanisms. Methods Liver cancer cell lines Huh7 and HepG2 stably transfected with HBV DNA polymerase expression plasmid with Flag (Flag-HBV-P) and intercellular adhesion molecule-1 (ICAM1) were co-cultured with Jurkat cells, and MTT assay, qRT-PCR, and ELISA were used to measure Jurkat cell proliferation, activation (CD69 expression), and secretion of the cytokine IFN-γ. RNA-seq was used to screen for differentially expressed immune-associated molecules between stably transfected cell lines and control cells, and mRNA half-life and protein half-life assays were used to determine the specific levels of the immune-associated molecules that were affected by HBV DNA polymerase. Related websites were used to predict the transcription factors that may bind to the promoter region of this immune-associated molecule, Western blot was used to verify the effect of transcription factors on the immune-associated molecule, and rescue experiment was used to determine whether HBV DNA polymerase affects the expression level of the immune-associated molecule through this transcription factor. The independent-samples t test was used for comparison between two groups. Results The experimental group had significant reductions in Jurkat cell proliferation, activation, and cytokine secretion compared with the control group (all P<0.01). Compared with the control group, the experimental group (Huh7 and HepG2 cell lines) had significant reductions in the mRNA and protein expression levels of ICAM1 (all P<0.01). Website prediction identified the ICAM1 promoter and preliminarily highlighted NFKB1, RELA, and STAT3. Compared with the control group, the experimental group (Huh7 and HepG2 cell lines) had a significant reduction in the protein expression level of p65 (all P<0.01). After p65 overexpression, there was a significant increase in the protein expression level of ICAM1, and after the expression of p65 was reduced, there was a significant reduction in the protein expression level of ICAM1 (all P<0.01). In the rescue experiment, there was no significant difference in the protein expression level of ICAM1 between the control group and the experimental group after p65 overexpression (all P>0.05). After the overexpression of ICAM1, there were no significant differences in the proliferation, activation, and cytokine secretion of Jurkat cells between the control group and the experimental group (Huh7 and HepG2 cell lines) (all P>0.05). Conclusion HBV DNA polymerase downregulates the level of ICAM1 to mediate HCC immune escape by inhibiting the expression of p65 in NF-κB. -
表 1 引物序列
Table 1. Primer Sequences
基因 上游引物(5′-3′) 下游引物(5′-3′) Flag-HBV-P GCCGGTACCATGCCCCTATCCTATCAACA ATAAGAATGCGGCCGCCGGTGGTCTCCATGCGAC pcDNA3-ICAM1 CTAGAATTCATGGCTCCCAGCAGCCCCCG GCTCTAGAGGGAGGCGTGGCTTGTGTGT pcDNA3-p65 CCCAAGCTTATGGACGAACTGTTC GCTCTAGAGGAGCTGATCTGACT shR-p65 ATAGGATCCCCGGAGAAACGTAAAAGGACATTCAAGAGATGTCCTTTTACGTTTCTCCTTTTTAAGCTTATA TATAAGCTTAAAAAGGAGAAACGTAAAAGGACATCTCTTGAATGTCCTTTTACGTTTCTCCGGGGATCCTAT -
[1] MODY K, ABOU-ALFA GK. Systemic therapy for advanced hepatocellular carcinoma in an evolving landscape[J]. Curr Treat Options Oncol, 2019, 20( 2): 3. DOI: 10.1007/s11864-019-0601-1. [2] BRAY F, FERLAY J, SOERJOMATARAM I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68( 6): 394- 424. DOI: 10.3322/caac.21492. [3] SCHLABE S, ROCKSTROH JK. Advances in the treatment of HIV/HCV coinfection in adults[J]. Expert Opin Pharmacother, 2018, 19( 1): 49- 64. DOI: 10.1080/14656566.2017.1419185. [4] CHEN Z, XIE H, HU M, et al. Recent progress in treatment of hepatocellular carcinoma[J]. Am J Cancer Res, 2020, 10( 9): 2993- 3036. [5] NORDENSTEDT H, WHITE DL, EL-SERAG HB. The changing pattern of epidemiology in hepatocellular carcinoma[J]. Dig Liver Dis, 2010, 42( Suppl 3): S206- S214. DOI: 10.1016/S1590-8658(10)60507-5. [6] SCHWEITZER A, HORN J, MIKOLAJCZYK RT, et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013[J]. Lancet, 2015, 386( 10003): 1546- 1555. DOI: 10.1016/S0140-6736(15)61412-X. [7] WILD CP, MILLER JD, GROOPMAN JD, et al. Mycotoxin control in low-and middle-income countries[M]. Lyon(FR): International Agency for Research on Cancer © International Agency for Research on Cancer, 2015. [8] BARTENSCHLAGER R, SCHALLER H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome[J]. EMBO J, 1992, 11( 9): 3413- 3420. DOI: 10.1002/j.1460-2075.1992.tb05420.x. [9] HIRSCH RC, LOEB DD, POLLACK JR, et al. cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA[J]. J Virol, 1991, 65( 6): 3309- 3316. DOI: 10.1128/JVI.65.6.3309-3316.1991. [10] WEI L, PLOSS A. Hepatitis B virus cccDNA is formed through distinct repair processes of each strand[J]. Nat Commun, 2021, 12( 1): 1591. DOI: 10.1038/s41467-021-21850-9. [11] SELZER L, ZLOTNICK A. Assembly and release of hepatitis B virus[J]. Cold Spring Harb Perspect Med, 2015, 5( 12): a021394. DOI: 10.1101/cshperspect.a021394. [12] ZLOTNICK A, VENKATAKRISHNAN B, TAN Z, et al. Core protein: A pleiotropic keystone in the HBV lifecycle[J]. Antiviral Res, 2015, 121: 82- 93. DOI: 10.1016/j.antiviral.2015.06.020. [13] INOUE T, TANAKA Y. The Role of hepatitis B core-related antigen[J]. Genes(Basel), 2019, 10( 5): 357. DOI: 10.3390/genes10050357. [14] BOUCHARD MJ, SCHNEIDER RJ. The enigmatic X gene of hepatitis B virus[J]. J Virol, 2004, 78( 23): 12725- 12734. DOI: 10.1128/JVI.78.23.12725-12734.2004. [15] BENHENDA S, COUGOT D, BUENDIA MA, et al. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis[J]. Adv Cancer Res, 2009, 103: 75- 109. DOI: 10.1016/S0065-230X(09)03004-8. [16] SLAGLE BL, BOUCHARD MJ. Role of HBx in hepatitis B virus persistence and its therapeutic implications[J]. Curr Opin Virol, 2018, 30: 32- 38. DOI: 10.1016/j.coviro.2018.01.007. [17] BELLONI L, POLLICINO T, de NICOLA F, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function[J]. Proc Natl Acad Sci U S A, 2009, 106( 47): 19975- 19979. DOI: 10.1073/pnas.0908365106. [18] HU J, FLORES D, TOFT D, et al. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function[J]. J Virol, 2004, 78( 23): 13122- 13131. DOI: 10.1128/JVI.78.23.13122-13131.2004. [19] KIM S, WANG H, RYU WS. Incorporation of eukaryotic translation initiation factor eIF4E into viral nucleocapsids via interaction with hepatitis B virus polymerase[J]. J Virol, 2010, 84( 1): 52- 58. DOI: 10.1128/JVI.01232-09. [20] JONES SA, HU J. Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging targets for antiviral intervention[J]. Emerg Microbes Infect, 2013, 2( 9): e56. DOI: 10.1038/emi.2013.56. [21] YU S, CHEN J, WU M, et al. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKepsilon and DDX3[J]. J Gen Virol, 2010, 91( Pt 8): 2080- 2090. DOI: 10.1099/vir.0.020552-0. [22] WANG H, RYU WS. Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: implications for immune evasion[J]. PLoS Pathog, 2010, 6( 7): e1000986. DOI: 10.1371/journal.ppat.1000986. [23] LIU Y, LI J, CHEN J, et al. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways[J]. J Virol, 2015, 89( 4): 2287- 2300. DOI: 10.1128/JVI.02760-14. [24] SUN Y, YU M, QU M, et al. Hepatitis B virus-triggered PTEN/β-catenin/c-Myc signaling enhances PD-L1 expression to promote immune evasion[J]. Am J Physiol Gastrointest Liver Physiol, 2020, 318( 1): G162- G173. DOI: 10.1152/ajpgi.00197.2019. [25] LIU M, WU H, LIU T, et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma[J]. Cell Res, 2009, 19( 7): 828- 837. DOI: 10.1038/cr.2009.72. [26] CHEN Y, XU Y, ZHAO M, et al. High-throughput T cell receptor sequencing reveals distinct repertoires between tumor and adjacent non-tumor tissues in HBV-associated HCC[J]. Oncoimmunology, 2016, 5( 10): e1219010. DOI: 10.1080/2162402X.2016.1219010. [27] STAUNTON DE, MARLIN SD, STRATOWA C, et al. Primary structure of ICAM-1 demonstrates interaction between members of the immunoglobulin and integrin supergene families[J]. Cell, 1988, 52( 6): 925- 933. DOI: 10.1016/0092-8674(88)90434-5. [28] HUBBARD AK, ROTHLEIN R. Intercellular adhesion molecule-1(ICAM-1) expression and cell signaling cascades[J]. Free Radic Biol Med, 2000, 28( 9): 1379- 1386. DOI: 10.1016/s0891-5849(00)00223-9. [29] WEE H, OH HM, JO JH, et al. ICAM-1/LFA-1 interaction contributes to the induction of endothelial cell-cell separation: implication for enhanced leukocyte diapedesis[J]. Exp Mol Med, 2009, 41( 5): 341- 348. DOI: 10.3858/emm.2009.41.5.038. [30] GORINA R, LYCK R, VESTWEBER D, et al. β2 integrin-mediated crawling on endothelial ICAM-1 and ICAM-2 is a prerequisite for transcellular neutrophil diapedesis across the inflamed blood-brain barrier[J]. J Immunol, 2014, 192( 1): 324- 337. DOI: 10.4049/jimmunol.1300858. [31] BACHMANN MF, MCKALL-FAIENZA K, SCHMITS R, et al. Distinct roles for LFA-1 and CD28 during activation of naive T cells: adhesion versus costimulation[J]. Immunity, 1997, 7( 4): 549- 557. DOI: 10.1016/s1074-7613(00)80376-3. [32] JENKINSON SR, WILLIAMS NA, MORGAN DJ. The role of intercellular adhesion molecule-1/LFA-1 interactions in the generation of tumor-specific CD8+ T cell responses[J]. J Immunol, 2005, 174( 6): 3401- 3407. DOI: 10.4049/jimmunol.174.6.3401. [33] POGGI A, PREVOSTO C, ZANCOLLI M, et al. NKG2D and natural cytotoxicity receptors are involved in natural killer cell interaction with self-antigen presenting cells and stromal cells[J]. Ann N Y Acad Sci, 2007, 1109: 47- 57. DOI: 10.1196/annals.1398.007. -



 PDF下载 ( 2211 KB)
PDF下载 ( 2211 KB)


 下载:
下载: