Pnpla3 I148M和Tm6sf2 E167K双突变纯合小鼠模型的构建
DOI: 10.3969/j.issn.1001-5256.2022.08.013
-
摘要:
目的 利用Pnpla3148M/M纯合小鼠与Tm6sf2167K/K纯合小鼠杂交方法构建Pnpla3148M/M Tm6sf2167K/K双突变小鼠模型。 方法 利用Pnpla3 Ⅰ 148M和Tm6sf2 E 167K单突变纯合小鼠交配出Pnpla3148M/M Tm6sf2167K/K双突变杂合小鼠,再通过自交得到Pnpla3148M/M Tm6sf2167K/K双突变纯合小鼠。选取同窝的Pnpla3148M/M Tm6sf2167K/K(n=6)、Pnpla3148M/M Tm6sf2167E/E(n=6)、Pnpla3148I/I Tm6sf2167K/K(n=6)和野生(Wt)(n=6)雄性小鼠普通饮食喂养8周,在第8周检测小鼠葡萄糖及脂质代谢等指标。多组间比较采用方差分析,进一步两两比较采用LSD-t检验。 结果 琼脂糖凝胶电泳和核酸测序结果表明Pnpla3148M/M Tm6sf2167K/K双突变小鼠模型构建成功。Pnpla3148M/M Tm6sf2167K/K小鼠体质量与其他3种基因型小鼠比较,差异无统计学意义(P值均>0.05),Pnpla3148M/M Tm6sf2167K/K小鼠肝湿重高于Wt小鼠(P<0.05)。Pnpla3148M/M Tm6sf2167K/K小鼠的空腹血糖低于Tm6sf2167K/K单突变小鼠和Wt小鼠(P值均<0.05),Pnpla3148M/M Tm6sf2167K/K小鼠葡萄糖耐受能力较Tm6sf2167K/K单突变小鼠有所下降(P<0.05),四组基因型小鼠的胰岛素水平无明显差异(P值均>0.05)。Pnpla3148M/M Tm6sf2167K/K双突变小鼠的血浆生化指标与其他三种基因型小鼠比较,差异均无统计学意义(P值均>0.05)。肝脏油红O切片染色结果显示Pnpla3148M/M Tm6sf2167K/K双突变小鼠的肝脏较Pnpla3148M/M单突变小鼠和Wt小鼠更容易发生脂质积累。 结论 Pnpla3148M/M Tm6sf2167K/K双突变小鼠模型构建成功,Pnpla3 Ⅰ 148M和Tm6sf2 E 167K双突变可引起小鼠葡萄糖代谢异常。 Abstract:Objective To construct a Pnpla3148M/M Tm6sf2167K/K double mutant mouse model by crossbreeding Pnpla3148M/M homozygous mice and Tm6sf2167K/K homozygous mice. Methods Pnpla3148I/M Tm6sf2167E/K heterozygous mice were bred by hybridization of Pnpla3148M/M Tm6sf2167E/E and Pnpla3148I/I Tm6sf2167K/K homozygous mice, and the Pnpla3148M/M Tm6sf2167K/K mice were obtained by the self-crossbreeding of Pnpla3148I/M Tm6sf2167E/K mice. Male mice of Pnpla3148M/M Tm6sf2167K/K (n=6), Pnpla3148M/M Tm6sf2167E/E (n=6), and Pnpla3148I/I Tm6sf2167K/K (n=6) genotypes and Wt mice (n=6) were fed with normal diet for 8 weeks, and then the glucose and lipid metabolism indices were measured. A one-way analysis of variance was used for comparison between multiple groups, and the least significant difference t-test was used for further comparison bewteen two groups. Results Agarose gel electrophoresis and nucleic acid sequencing results showed that the Pnpla3148M/M Tm6sf2167K/K double mutant mouse model was successfully constructed. There were no significant difference in body weight between the Pnpla3148M/M Tm6sf2167K/K mice and the Pnpla3148M/M Tm6sf2167E/E, Pnpla3148I/I Tm6sf2167K/K, and Wt mice (all P > 0.05). The Pnpla3148M/M Tm6sf2167K/K mice had a significantly higher liver wet weight than the Wt mice (P < 0.05). The fasting blood glucose of Pnpla3148M/M Tm6sf2167K/K mice was significantly lower than that of Pnpla3148I/I Tm6sf2167K/K mice and Wt mice (both P < 0.05). The glucose tolerance of Pnpla3148M/M Tm6sf2167K/K mice was significantly reduced compared with the Pnpla3148I/I Tm6sf2167K/K mice (P < 0.05). There were no significant differences in insulin level between the four groups of mice (all P > 0.05). Also, there were no significant differences in the serum levels of biochemical indices between the Pnpla3148M/M Tm6sf2167K/K mice and the Pnpla3148M/M Tm6sf2167E/E, Pnpla3148I/I Tm6sf2167K/K, and Wt mice (all P > 0.05). Oil red O staining of the liver showed that more lipid accumulation was observed in the Pnpla3148M/M Tm6sf2167K/K mice than in the Pnpla3148M/M Tm6sf2167E/E and Wt mice. Conclusion The Pnpla3148M/M Tm6sf2167K/K double mutant mouse model was successfully constructed. Pnpla3 Ⅰ 148M and Tm6sf2 E 167K double mutations can cause abnormal glucose metabolism in mice. -
Key words:
- Non-alcoholic Fatty Liver Disease /
- Point Mutation /
- Models, Animal /
- Mice
-
非酒精性脂肪性肝病(NAFLD)是指除酒精和其他明确的损肝因素所致的、以弥漫性肝细胞大泡性脂肪变性为主要特征的临床病理综合征[1-3]。有研究[4-6]发现NAFLD与胰岛素抵抗和遗传易感性显著相关。含patatin样磷脂酶域3(patatin-like phospholipase domain containing 3, Pnpla3)位于22号染色体上[7], 主要表达于脂肪组织和肝脏[8]。跨膜蛋白6超家族成员2基因(transmembrane 6 superfamily member 2,Tm6sf2)位于第19号染色体上,TM6SF2主要定位于内质网膜,主要表达于肝脏和小肠[9],这两个器官参与全身脂质稳态的调节[10]。全基因组关联和外显子组测序结果显示Pnpla3 I148M和Tm6sf2 E167K基因的遗传变异与NAFLD的易感性和进展相关,Pnpla3 I148M是指该蛋白第148位异亮氨酸被蛋氨酸取代,Tm6sf2 E167K是指该蛋白第167位谷氨酸被赖氨酸取代,Pnpla3和Tm6sf2基因的遗传变异与肝脂肪含量增加、进展为非酒精性脂肪型肝炎、肝硬化和肝细胞癌密切相关[11-13]。人群研究[14-15]发现Pnpla3 I148M和Tm6sf2 E167K双突变对NAFLD的发展产生叠加效应,协同参与肝脏脂肪变性的发展。但是Pnpla3 I148M联合Tm6sf2 E167K对NAFLD的共同作用及机理尚不明确,因此本研究旨在构建Pnpla3 I148M和Tm6sf2 E167K双突变小鼠模型,探究其在常规饮食条件下体内脂质代谢的变化,为探究Pnpla3 I148M和Tm6sf2 E167K双突变在NAFLD发病中的联合作用及相关机制提供良好的动物模型。
1. 材料与方法
1.1 材料
Tm6sf2 E167K小鼠与Pnpla3 I148M小鼠均为本课题组所有(青岛大学附属青岛市市立医院肝病研究室),所有小鼠均饲养于SPF级动物房。实验动物生产许可证: SCXK(苏)2018-0008,实验动物使用许可证: SYXK(鲁)20200009。TC、TG、AST和ALT检测试剂盒购买自南京建成生物工程研究所;血糖仪和血糖试纸购买自日本OMRON公司;胰岛素检测试剂盒购买自上海酶联生物科技有限公司。
1.2 Pnpla3148M/M Tm6sf2167K/K小鼠的繁育策略
Pnpla3148I/M和Tm6sf2167E/K杂合小鼠通过各自自交,分别得到Pnpla3148M/M和Tm6sf2167K/K单突变纯合小鼠,通过Pnpla3148M/M单突变纯合小鼠与Tm6sf2167K/K单突变纯合小鼠交配,得到Pnpla3148I/M Tm6sf2167E/K双突变杂合小鼠,最后通过Pnpla3148I/M Tm6sf2167E/K双突变杂合小鼠自交,得到Pnpla3148M/M Tm6sf2167K/K双突变纯合小鼠(图 1)。统计Pnpla3148I/M Tm6sf2167E/K双突变杂合小鼠自交后的子代中Pnpla3148M/M Tm6sf2167K/K小鼠、Pnpla3148M/M单突变纯合小鼠、Tm6sf2167K/K单突变纯合小鼠、杂合小鼠(包括Pnpla3 I148M或Tm6sf2 E167K单突变杂合小鼠及双突变杂合小鼠)和Wt小鼠的数量,计算各种基因型小鼠数量占总体的比例,检验是否符合孟德尔遗传分离定律。
1.3 Pnpla3148M/M Tm6sf2167K/K小鼠的基因鉴定
剪取1~2周龄小鼠0.3 cm小鼠尾尖于1.5 mL EP管中,加入60 μL碱性裂解液A(A液:25 mmol/L氢氧化钠;20 μmol/L乙二胺四乙酸二钠),95 ℃金属浴40 min后加入60 μL裂解液B(B液:40 mmol/L Tris-Cl), 机械研磨3 min,6000 r/min离心6 min得到上清液即为粗提所得DNA, 以其作为模板进行PCR扩增。将PCR扩增产物进行琼脂糖凝胶电泳,根据条带大小来鉴定Tm6sf2基因型,利用基因测序方法鉴定Pnpla3基因型。Tm6sf2和Pnpla3的PCR引物见表 1,Pnpla3测序引物见表 2。
表 1 小鼠基因型鉴定引物序列Table 1. Primer sequences for mouse genotypic identificationPCR引物名称 引物序列(5′-3′) Pnpla3-wt-tF1 ATCTCTGTGAGTTCGATTGCCAG Pnpla3-wt-tR1 AGTGTATCCAACAGACAGCAGGC Tm6sf2-wt-tF1 GGCCTTTCCTAGACTCCTCA Tm6sf2-wt-tR1 CCTTCTCAGATGTTCCTCCCT 表 2 Pnpla3小鼠基因型测序引物序列Table 2. Primer sequences for Pnpla3 mouse genotype sequencingPCR引物名称 引物序列(5′-3′) Pnpla3-wt-tR1 AGTGTATCCAACAGACAGCAGGC 1.4 小鼠口服普通糖耐量试验(oral glucose tolerance test,OGTT)
选择8周龄同窝别Pnpla3148M/M Tm6sf2167K/K(n=6)、Pnpla3148M/M Tm6sf2167E/E(n=6)、Pnpla3148I/I Tm6sf2167K/K(n=6) 和Wt(n=6)雄性小鼠,提前数天给小鼠做掐尾训练。实验当天于早上8点给予小鼠禁食6 h,禁食结束后称量体质量并检测鼠尾静脉空腹血糖,然后给每只小鼠进行葡萄糖灌胃(2 g葡萄糖/kg体质量)[16]。分别测量灌胃后30、60、120 min时的小鼠尾静脉血糖,记录血糖值并制作OGTT曲线。
1.5 小鼠生化指标检测
选择8周龄同窝别Pnpla3148M/M Tm6sf2167K/K(n=6)、Pnpla3148M/M Tm6sf2167E/E(n=6)、Pnpla3148I/I Tm6sf2167K/K(n=6) 和Wt(n=6)雄性小鼠,禁食禁水6 h后,称量体质量,利用4%水合氯醛对小鼠进行麻醉后取血(采用乙二胺四乙酸二钠抗凝),2500×g,4 ℃离心20 min,取上层血浆。然后解剖出小鼠的主要组织器官进行形态学对比,并计算肝脏指数。按照说明书方法测量小鼠血浆TC、TG、AST、ALT和胰岛素水平。
1.6 Pnpla3148M/M Tm6sf2167K/K小鼠肝组织学评估
取上述8周龄雄性小鼠的肝脏,用0.9%等渗生理盐水清洗后用4%多聚甲醛固定,脱水后用石蜡包埋并切片,分别做小鼠肝组织的HE染色和油红O染色,光学显微镜观察并拍照。
1.7 统计学方法
数据使用GraphPad prism 8软件处理,多组间比较采用方差分析,进一步两两比较采用LSD-t检验。P<0.05为差异有统计学意义。
2. 结果
2.1 Pnpla3148M/M Tm6sf2167K/K小鼠的基因鉴定
取小鼠鼠尾DNA进行PCR扩增,扩增后将产物进行1.5%琼脂糖凝胶电泳。Tm6sf2基因型的鉴定结果为:琼脂糖凝胶电泳结果只有一条203 bp条带则是Tm6sf2167K/K纯合突变小鼠,只有一条147 bp条带则是Tm6sf2167E/E野生小鼠,同时扩增出上述两条条带则是Tm6sf2167E/K杂合小鼠。Pnpla3基因型的鉴定结果为:琼脂糖凝胶电泳显示扩增出506 bp条带后进行基因测序,测序结果证实第444位密码子由ATT突变为ATG,为Pnpla3148M/M小鼠,若同时存在ATT和ATG两种碱基类型,则为Pnpla3148I/M小鼠(图 2)。
2.2 孟德尔遗传分离定律检测
将Pnpla3148I/M Tm6sf2167E/K小鼠进行自交,统计子代Pnpla3148M/M Tm6sf2167K/K、Pnpla3148M/M Tm6sf2167E/E、Pnpla3148I/I Tm6sf2167K/K、杂合(Pnpla3148I/M Tm6sf2167E/K、Pnpla3148I/M Tm6sf2167E/E、Pnpla3148I/I Tm6sf2167E/K、Pnpla3148M/M Tm6sf2167E/K和Pnpla3148I/M Tm6sf2167K/K)和Wt小鼠的数量,结果见表 3,各种基因型小鼠数量总体比例大致为1∶ 1∶ 1∶ 12∶ 1,符合孟德尔遗传分离定律,并且子代小鼠生长状况正常,无死亡。这些结果说明Pnpla3148I/M Tm6sf2167E/K双突变小鼠能够进行正常繁育后代,并且后代可以正常存活。
表 3 Pnpla3148I/M Tm6sf2167E/K小鼠自交后子代各基因型数量Table 3. Number of progenies with each genotype after Pnpla3148I/ MTM6SF2167E/K mice self-breeding小鼠基因型 数量 占总体的百分比(%) Pnpla3148M/M Tm6sf2167K/K 13 6.2 杂合小鼠 151 71.9 Pnpla3148M/M Tm6sf2167E/E 11 5.2 Pnpla3148I/I Tm6sf2167K/K 18 8.6 Wt小鼠 17 8.1 2.3 不同基因型小鼠体质量和肝指数的差异检测
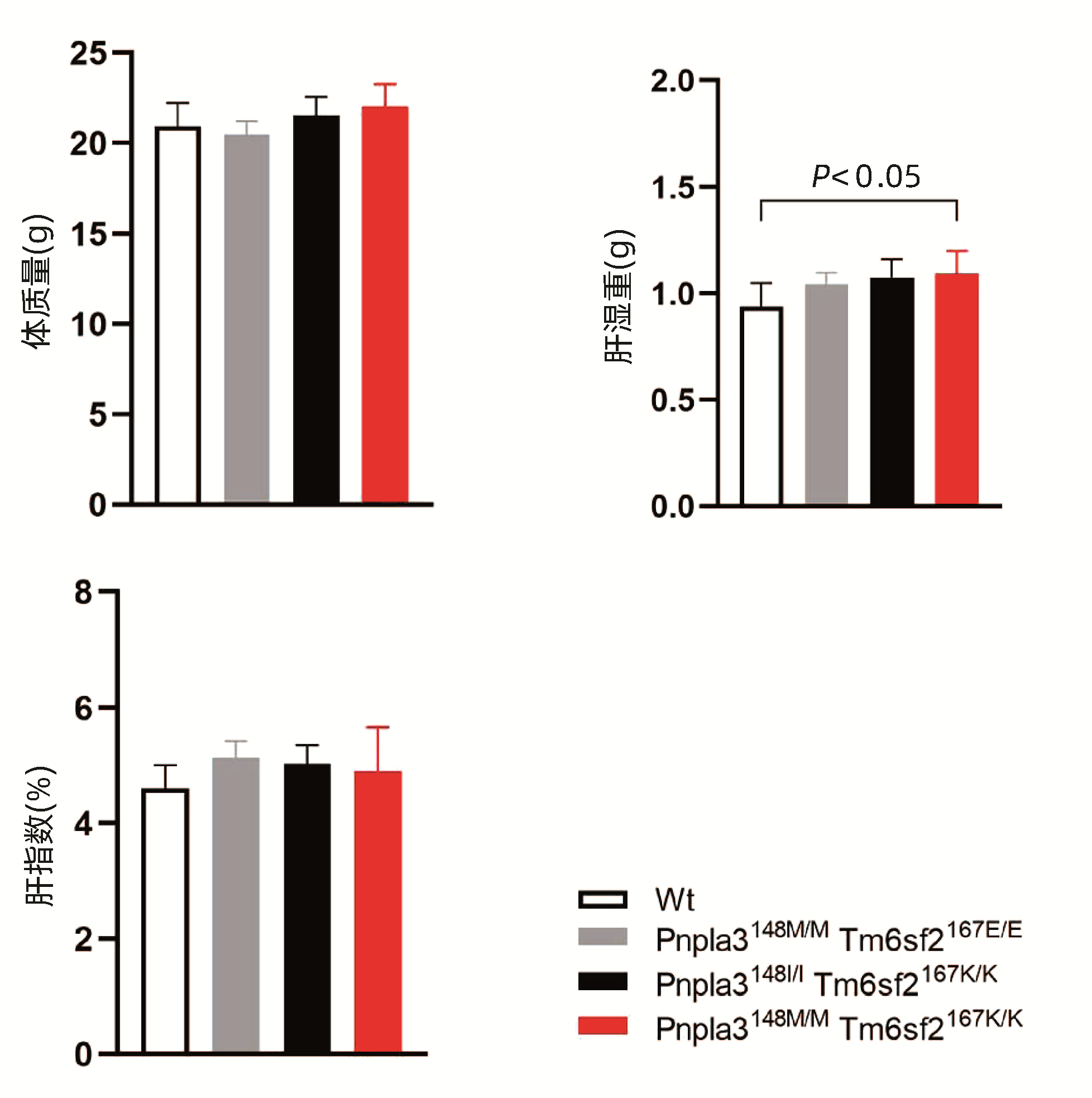
Pnpla3148M/M Tm6sf2167K/K、Pnpla3148M/M Tm6sf2167E/E、Pnpla3148I/I Tm6sf2167K/K和Wt小鼠以普通饮食喂养8周,检测不同基因型小鼠的体质量变化。结果显示,Pnpla3148M/M Tm6sf2167K/K基因型小鼠与其他三种基因型小鼠体质量无明显差异(P值均>0.05),检测小鼠肝湿重发现Pnpla3148M/M Tm6sf2167K/K小鼠高于Wt小鼠的肝湿重(P<0.05),但通过计算各组小鼠的肝指数发现,Pnpla3148M/M Tm6sf2167K/K基因型小鼠的肝指数与其他三种基因型小鼠的肝指数比较,差异无统计学意义(P值均>0.05)(图 3)。
2.4 不同基因型小鼠各组织的表型比较
分别解剖8周龄的Pnpla3148M/M Tm6sf2167K/K、Pnpla3148M/M Tm6sf2167E/E、Pnpla3148I/I Tm6sf2167K/K和Wt小鼠的脑、甲状腺、心脏、肺、肝脏、脾脏、胃、肠、肾和睾丸,对比四组小鼠各个器官的大小和形态,发现四组小鼠之间的器官形态无明显差异(图 4)。
2.5 不同基因型小鼠的葡萄糖代谢差异
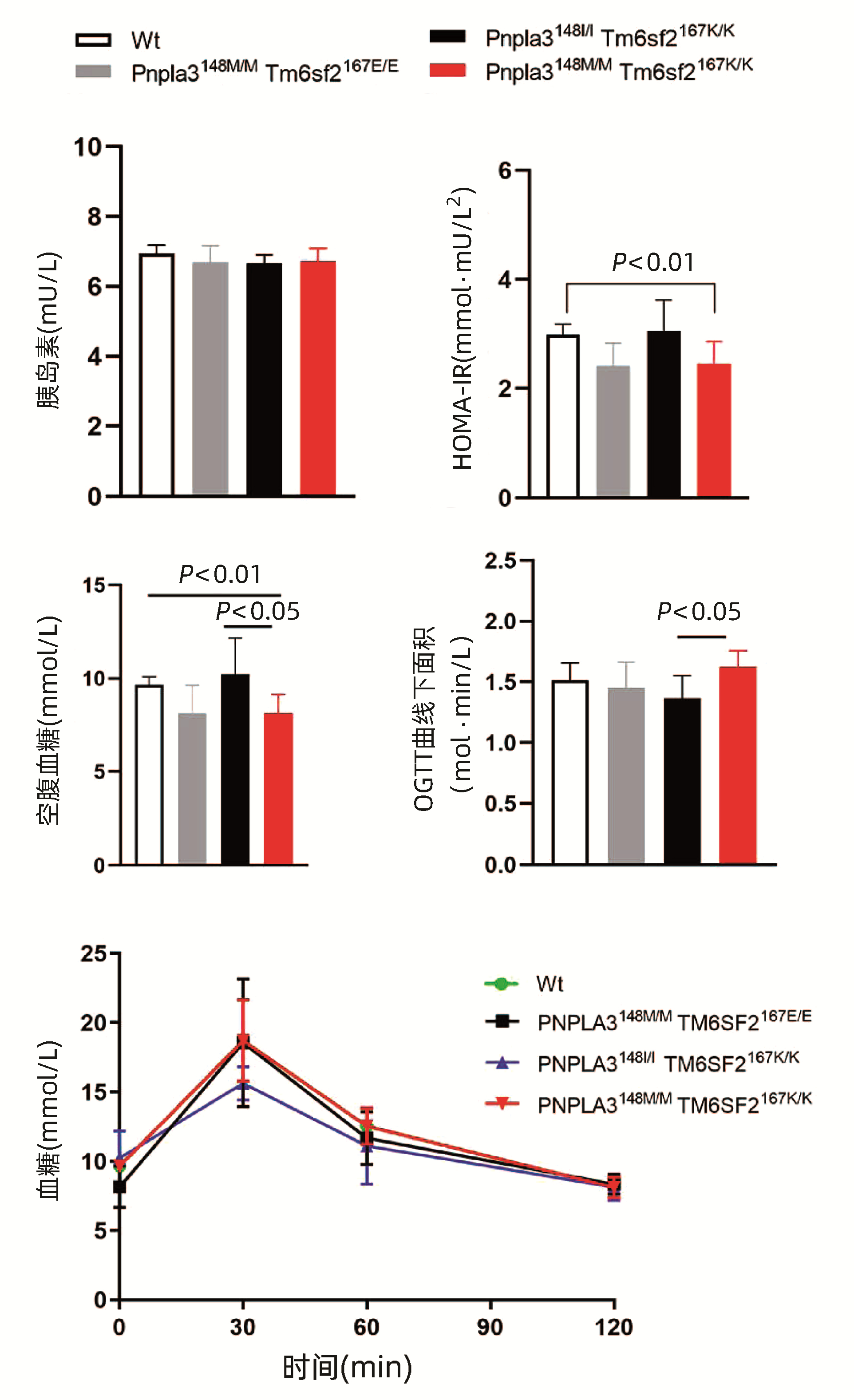
对8周龄的Pnpla3148M/M Tm6sf2167K/K、Pnpla3148M/M Tm6sf2167E/E、Pnpla3148I/I Tm6sf2167K/K和Wt雄性小鼠的空腹血糖进行检测,结果显示Pnpla3148M/M Tm6sf2167K/K空腹血糖低于Tm6sf2167K/K单突变小鼠和Wt小鼠,差异均有统计学意义(P值均<0.05),而与Pnpla3148M/M单突变小鼠无明显差异(P>0.05),提示Pnpla3 I148M突变会导致小鼠空腹血糖下降。OGTT曲线下面积比较结果显示,Pnpla3148M/M Tm6sf2167K/K小鼠OGTT曲线下面积较Tm6sf2167K/K单突变小鼠升高,差异有统计学意义(P<0.05),较Pnpla3148M/M单突变小鼠和Wt小鼠无明显差异(P值均>0.05),提示Pnpla3148M/M Tm6sf2167K/K双突变较Tm6sf2167K/K单突变更容易导致小鼠葡萄糖耐受能力的下降;对四组小鼠的胰岛素水平检测结果显示,胰岛素水平在不同基因型小鼠中的水平无明显差异(P值均>0.05)。Pnpla3148M/M Tm6sf2167K/K小鼠的胰岛素抵抗指数(HOMA-IR)较Wt小鼠有所降低,这与两组小鼠之间的空腹血糖含量趋势一致(图 5)。
2.6 不同基因型小鼠血浆生化指标检测
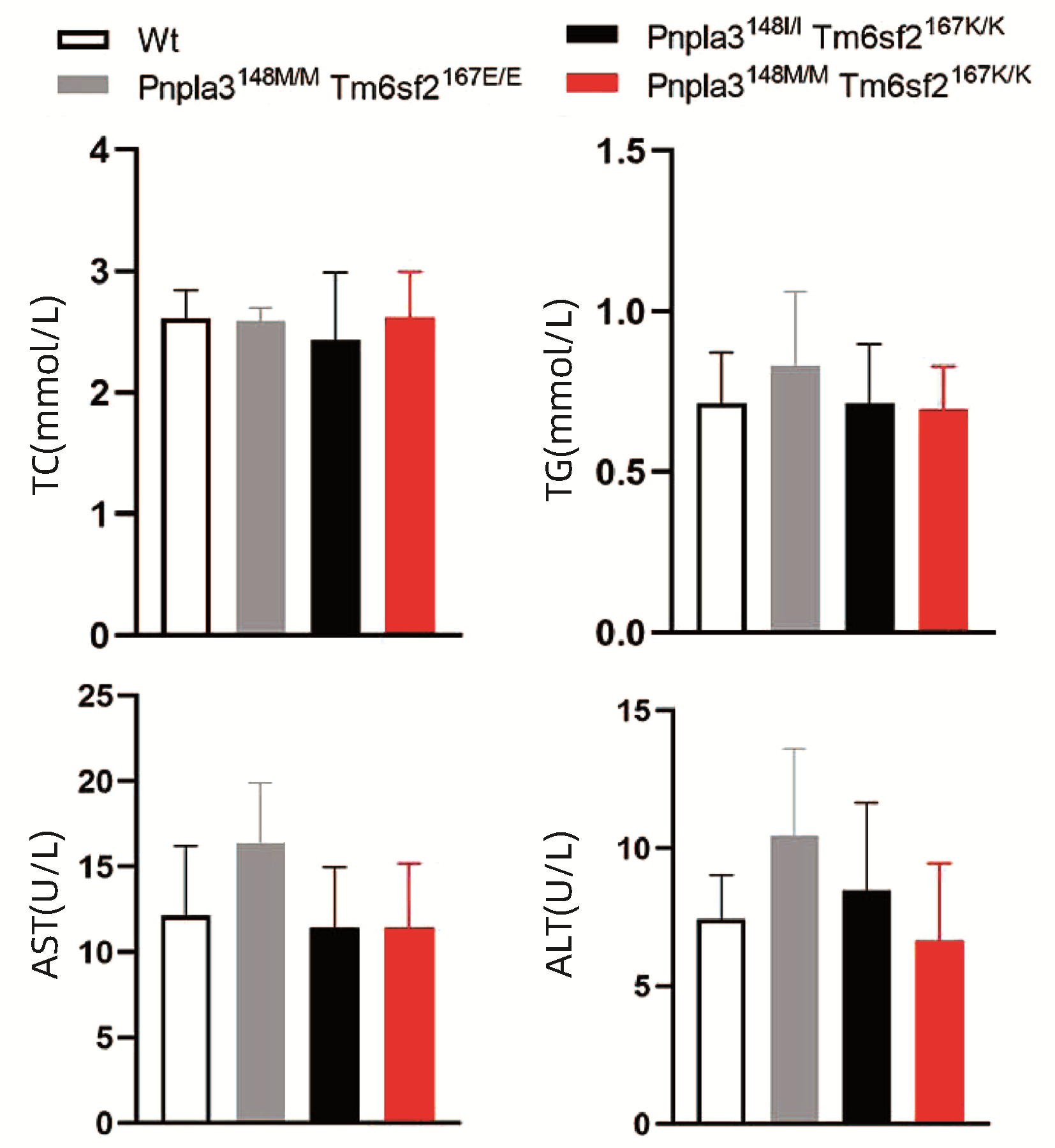
对8周龄Pnpla3148M/M Tm6sf2167K/K、Pnpla3148M/M Tm6sf2167E/E、Pnpla3148I/I Tm6sf2167K/K和Wt雄性小鼠的血浆TG、TC、AST和ALT水平进行检测,结果显示Pnpla3148M/M Tm6sf2167K/K小鼠血浆中的TG、TC、ALT和AST水平和另外三种基因型小鼠比较,差异均无统计学意义(P值均>0.05)(图 6)。
2.7 不同基因型小鼠肝脏HE染色和油红O染色结果
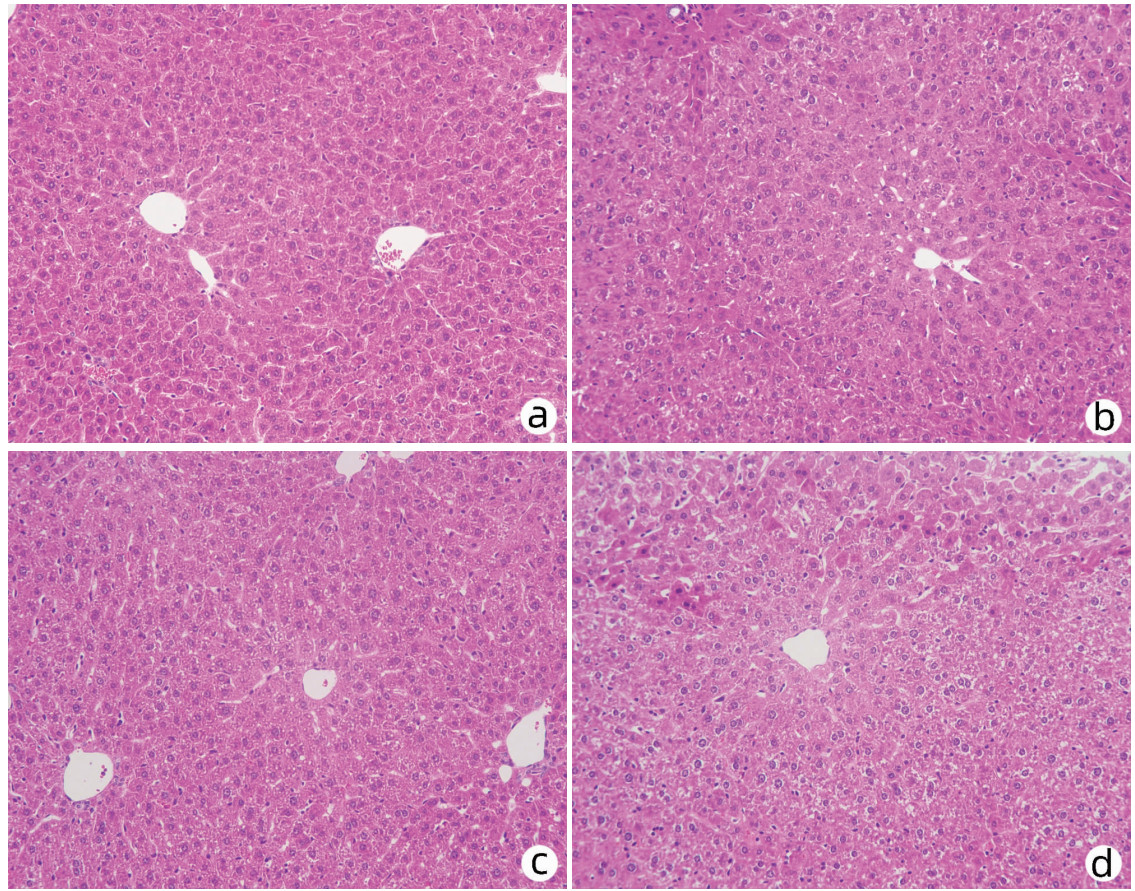
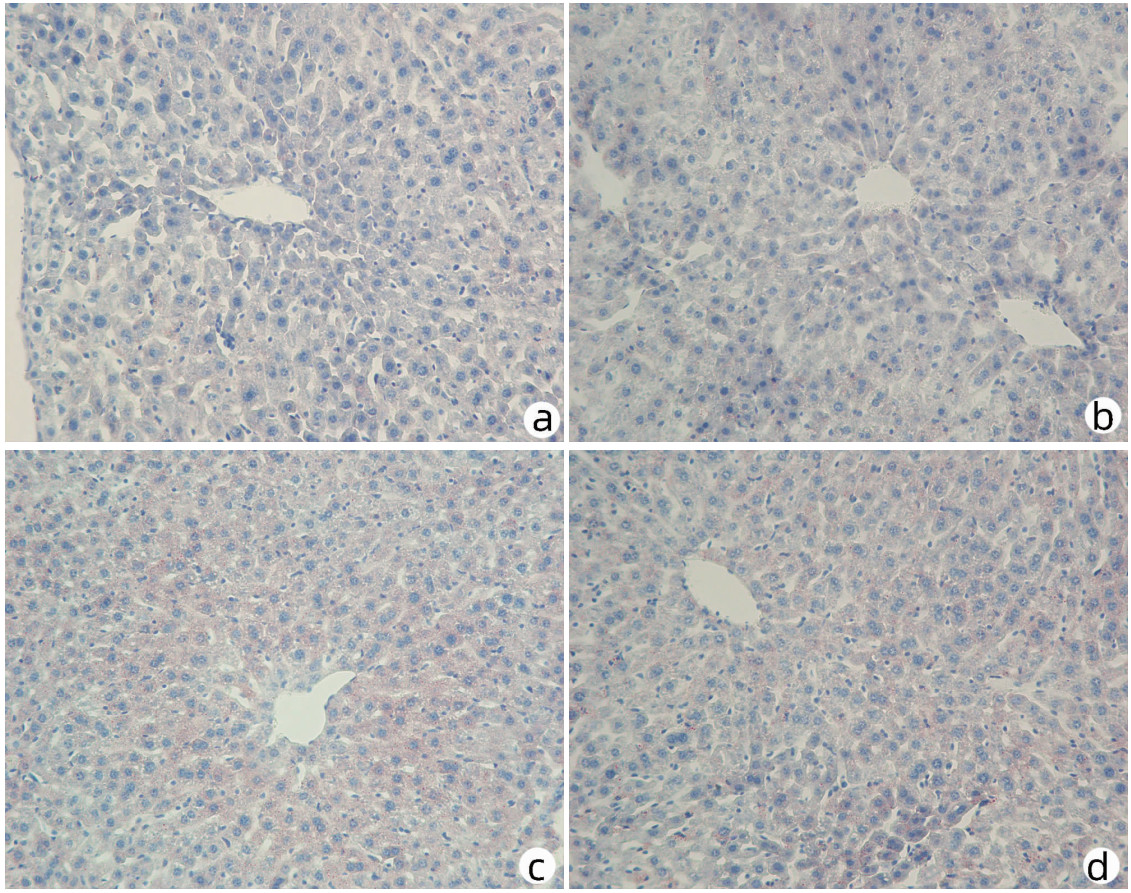
对8周龄Pnpla3148M/M Tm6sf2167K/K、Pnpla3148M/M Tm6sf2167E/E、Pnpla3148I/I Tm6sf2167K/K和Wt雄性小鼠的肝脏进行HE染色和油红O染色,结果显示Pnpla3148M/M Tm6sf2167K/K小鼠与Pnpla3 I148M、Tm6sf2 E167K单突变和Wt小鼠肝组织学之间无明显差异(图 7);油红O染色结果表明Pnpla3148M/M Tm6sf2167K/K小鼠比Pnpla3148M/M单突变小鼠和Wt小鼠肝脏内脂滴水平有所增多,提示Pnpla3 I148M和Tm6sf2 E167K双突变较Pnpla3 I148M单突变小鼠和Wt小鼠的肝脏更易发生脂质积累,但同Tm6sf2 E167K单突变小鼠相比无明显差异(图 8)。
3. 讨论
NAFLD患病人数的逐年增加给社会带来了沉重的医疗和经济负担,在全球范围内,NAFLD的平均发病率为25%[17]。尽管目前关于NAFLD的研究已经取得了显著进展,但是其发病机制至今尚未完全阐明。前期笔者的研究发现,Pnpla3 I148M和Tm6sf2 E167K在细胞水平上可发挥联合作用从而共同调节细胞内的脂质代谢过程[18],但未在动物水平进行验证。因此本文通过杂交繁育获得了Pnpla3148M/M Tm6sf2167K/K双突变小鼠,为进一步在动物水平探究PNPLA3 I148M及TM6SF2 E167K双突变引发肝脏脂肪变性的作用及分子机制奠定动物模型基础。
在本研究中,完成Pnpla3148M/M Tm6sf2167K/K双突变小鼠模型构建后,对Pnpla3148M/M Tm6sf2167K/K双突变小鼠的表型做了初步研究。本研究把PNPLA3 I148M和TM6SF2 E167K双突变杂合小鼠交配的后代分为Pnpla3148M/M Tm6sf2167K/K、Pnpla3148M/M Tm6sf2167E/E、Pnpla3148I/I Tm6sf2167K/K和Wt四组。统计Pnpla3148I/M Tm6sf2167E/K小鼠自交后代的存活数量,发现PNPLA3I148M和TM6SF2 E167K双突变小鼠能够正常存活,无胚胎致死情况发生。Pnpla3148M/M Tm6sf2167K/K小鼠的体质量较其他3种基因型小鼠体质量无明显差异,进一步对小鼠各器官形态进行观察发现,Pnpla3148M/M Tm6sf2167K/K小鼠各器官形态较其他3种基因型小鼠各器官形态无明显差异。检测小鼠葡萄糖代谢指标,糖耐量试验发现Pnpla3148M/M Tm6sf2167K/K和Pnpla3148M/M单突变小鼠空腹血糖无明显差异,但Pnpla3148M/M Tm6sf2167K/K小鼠的空腹血糖低于Tm6sf2167K/K单突变小鼠和Wt小鼠,提示Pnpla3 I148M和Tm6sf2 E167K双突变会降低小鼠的空腹血糖;OGTT曲线下面积是评价葡萄糖耐量的指标,结果显示Pnpla3148M/M Tm6sf2167K/K小鼠OGTT曲线下面积高于Tm6sf2167K/K单突变小鼠,表明Pnpla3148M/M Tm6sf2167K/K双突变小鼠的葡萄糖耐受能力较Tm6sf2167K/K单突变小鼠有所下降。
通过对小鼠血脂和肝功指标的检测发现,Pnpla3148M/M Tm6sf2167K/K双突变小鼠血浆中TC、TG、ALT和AST与其余三种基因型小鼠无明显差异。肝脏油红O染色提示Pnpla3148M/M Tm6sf2167K/K双突变较Pnpla3148M/M单突变小鼠和Wt小鼠更容易发生肝脏的脂质积累,但Pnpla3148M/M Tm6sf2167K/K双突变小鼠同Tm6sf2167K/K单突变小鼠的肝内脂滴水平无明显差异。
本研究成功构建了Pnpla3148M/M Tm6sf2167K/K双突变小鼠模型,该模型有利于在动物水平探究Pnpla3 I148M及Tm6sf2 E167K双突变引发肝脏脂肪变性的作用及分子机制,揭示Pnpla3 I148M和Tm6sf2 E167K双突变在NAFLD发病过程中的交互作用。
-
表 1 小鼠基因型鉴定引物序列
Table 1. Primer sequences for mouse genotypic identification
PCR引物名称 引物序列(5′-3′) Pnpla3-wt-tF1 ATCTCTGTGAGTTCGATTGCCAG Pnpla3-wt-tR1 AGTGTATCCAACAGACAGCAGGC Tm6sf2-wt-tF1 GGCCTTTCCTAGACTCCTCA Tm6sf2-wt-tR1 CCTTCTCAGATGTTCCTCCCT 表 2 Pnpla3小鼠基因型测序引物序列
Table 2. Primer sequences for Pnpla3 mouse genotype sequencing
PCR引物名称 引物序列(5′-3′) Pnpla3-wt-tR1 AGTGTATCCAACAGACAGCAGGC 表 3 Pnpla3148I/M Tm6sf2167E/K小鼠自交后子代各基因型数量
Table 3. Number of progenies with each genotype after Pnpla3148I/ MTM6SF2167E/K mice self-breeding
小鼠基因型 数量 占总体的百分比(%) Pnpla3148M/M Tm6sf2167K/K 13 6.2 杂合小鼠 151 71.9 Pnpla3148M/M Tm6sf2167E/E 11 5.2 Pnpla3148I/I Tm6sf2167K/K 18 8.6 Wt小鼠 17 8.1 -
[1] FRIEDMAN SL, NEUSCHWANDER-TETRI BA, RINELLA M, et al. Mechanisms of NAFLD development and therapeutic strategies[J]. Nat Med, 2018, 24(7): 908-922. DOI: 10.1038/s41591-018-0104-9. [2] YOUNOSSI Z, ANSTEE QM, MARIETTI M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention[J]. Nat Rev Gastroenterol Hepatol, 2018, 15(1): 11-20. DOI: 10.1038/nrgastro.2017.109. [3] ARON-WISNEWSKY J, VIGLIOTTI C, WITJES J, et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders[J]. Nat Rev Gastroenterol Hepatol, 2020, 17(5): 279-297. DOI: 10.1038/s41575-020-0269-9. [4] WATT MJ, MIOTTO PM, de NARDO W, et al. The liver as an endocrine organ-linking NAFLD and insulin resistance[J]. Endocr Rev, 2019, 40(5): 1367-1393. DOI: 10.1210/er.2019-00034. [5] POWELL EE, WONG VW, RINELLA M. Non-alcoholic fatty liver disease[J]. Lancet, 2021, 397(10290): 2212-2224. DOI: 10.1016/S0140-6736(20)32511-3. [6] TILG H, MOSCHEN AR, RODEN M. NAFLD and diabetes mellitus[J]. Nat Rev Gastroenterol Hepatol, 2017, 14(1): 32-42. DOI: 10.1038/nrgastro.2016.147. [7] TRÉPO E, ROMEO S, ZUCMAN-ROSSI J, et al. PNPLA3 gene in liver diseases[J]. J Hepatol, 2016, 65(2): 399-412. DOI: 10.1016/j.jhep.2016.03.011. [8] BASU RAY S. PNPLA3-I148M: a problem of plenty in non-alcoholic fatty liver disease[J]. Adipocyte, 2019, 8(1): 201-208. DOI: 10.1080/21623945.2019.1607423. [9] KOZLITINA J, SMAGRIS E, STENDER S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease[J]. Nat Genet, 2014, 46(4): 352-356. DOI: 10.1038/ng.2901. [10] O'HARE EA, YANG R, YERGES-ARMSTRONG LM, et al. TM6SF2 rs58542926 impacts lipid processing in liver and small intestine[J]. Hepatology, 2017, 65(5): 1526-1542. DOI: 10.1002/hep.29021. [11] STEFAN N, HÄRING HU, CUSI K. Non-alcoholic fatty liver disease: causes, diagnosis, cardiometabolic consequences, and treatment strategies[J]. Lancet Diabetes Endocrinol, 2019, 7(4): 313-324. DOI: 10.1016/S2213-8587(18)30154-2. [12] IOANNOU GN. Epidemiology and risk-stratification of NAFLD-associated HCC[J]. J Hepatol, 2021, 75(6): 1476-1484. DOI: 10.1016/j.jhep.2021.08.012. [13] LI JF, ZHENG EQ, XIE M. Association between rs738409 polymorphism in patatin-like phospholipase domain-containing protein 3 (PNPLA3) gene and hepatocellular carcinoma susceptibility: Evidence from case-control studies[J]. Gene, 2019, 685: 143-148. DOI: 10.1016/j.gene.2018.11.012. [14] WANG X, LIU Z, WANG K, et al. Additive effects of the risk alleles of PNPLA3 and TM6SF2 on non-alcoholic fatty liver disease (NAFLD) in a Chinese population[J]. Front Genet, 2016, 7: 140. DOI: 10.3389/fgene.2016.00140. [15] XU M, LI Y, ZHANG S, et al. Interaction of TM6SF2 E167K and PNPLA3 I148M variants in NAFLD in northeast China[J]. Ann Hepatol, 2019, 18(3): 456-460. DOI: 10.1016/j.aohep.2018.10.005. [16] ZHANG J, MA XF, WANG YF, et al. Hepatocyte-specific TM6SF2 knockout aggravates hepatic steatosis in mice with nonalcoholic fatty liver disease[J]. J Clin Hepatol, 2021, 37(11): 2612-2616. DOI: 10.3969/j.issn.1001-5256.2021.11.024.张杰, 马学峰, 王艺奋, 等. 肝脏TM6SF2特异性敲除促进非酒精性脂肪性肝病小鼠肝脏脂肪变性[J]. 临床肝胆病杂志, 2021, 37(11): 2612-2616. DOI: 10.3969/j.issn.1001-5256.2021.11.024. [17] LAZARUS JV, ANSTEE QM, HAGSTRÖM H, et al. Defining comprehensive models of care for NAFLD[J]. Nat Rev Gastroenterol Hepatol, 2021, 18(10): 717-729. DOI: 10.1038/s41575-021-00477-7. [18] CHEN L, DU S, LU L, et al. The additive effects of the TM6SF2 E167K and PNPLA3 I148M polymorphisms on lipid metabolism[J]. Oncotarget, 2017, 8(43): 74209-74216. DOI: 10.18632/oncotarget.18474. -




 PDF下载 ( 6045 KB)
PDF下载 ( 6045 KB)


 下载:
下载:








 下载:
下载: