基于影像组学与3D深度学习的多模态模型在重症急性胰腺炎预测中的应用
DOI: 10.12449/JCH251022
Application of a multimodal model based on radiomics and 3D deep learning in predicting severe acute pancreatitis
-
摘要:
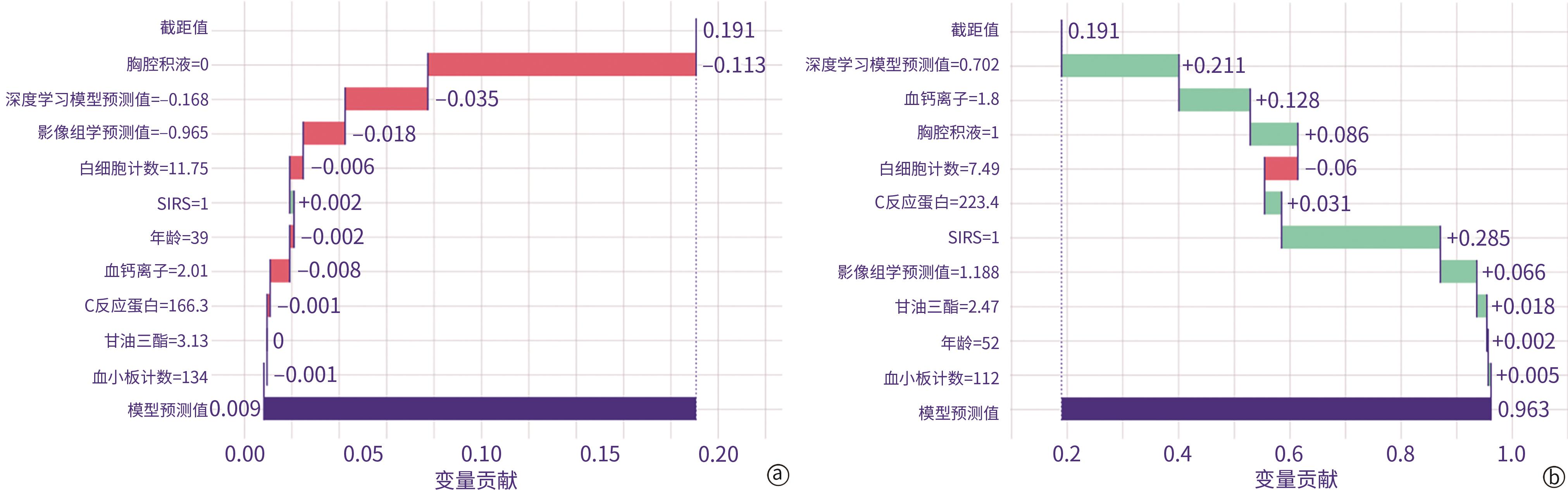
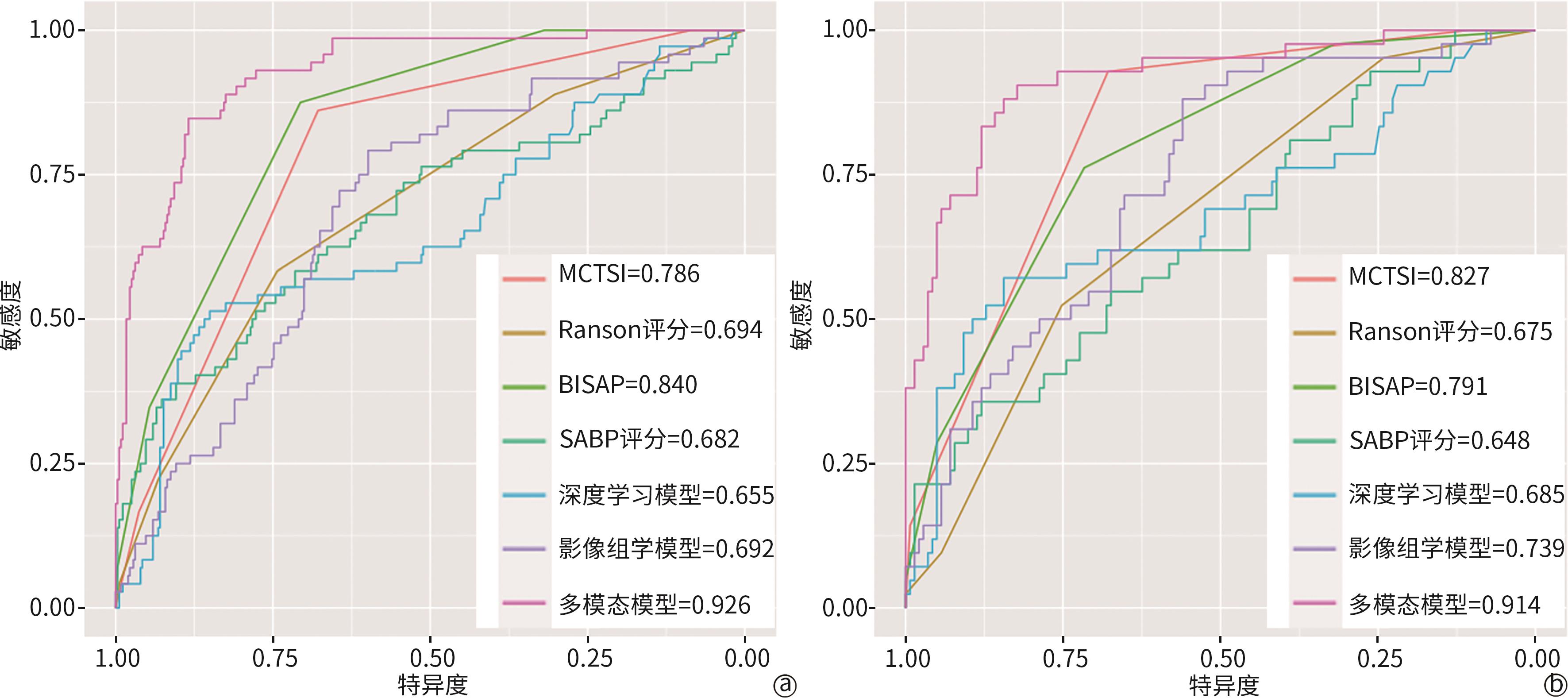
目的 探索融合影像组学特征、深度学习特征及临床结构化数据的多模态模型在重症急性胰腺炎(SAP)预测中的应用价值,以期为临床早期识别SAP提供更精准的工具。 方法 纳入2017年1月1日至2023年12月31日就诊于苏州大学附属第一医院、江苏大学附属金坛医院及苏州永鼎医院的急性胰腺炎(AP)患者,收集其人口学信息、既往史、病因、入院24 h内实验室检查数据及72 h内影像学资料,并评估全身炎症反应综合征(SIRS),同时计算Ranson评分、改良CT严重指数(MCTSI)、床边急性胰腺炎严重度指数(BISAP)和急性胰腺炎风险(SABP)评分。模型构建流程如下:(1)利用三维CT图像提取并筛选影像组学特征,基于极端梯度提升(XGBoost)算法建立影像组学分类模型;(2)采用U-Net对三维CT图像进行语义分割,随后将分割结果输入3D ResNet50构建深度学习分类模型;(3)基于XGBoost算法融合上述2种模型的预测值与临床结构化数据,建立多模态模型。采用变量重要性排序图和局部可解释性图对模型进行可视化解释。符合正态分布的计量资料组间比较采用成组t检验;不符合正态分布的采用Mann-Whitney U检验。计数资料组间比较采用χ2检验或Fisher精确检验。绘制各模型和已有评分系统的受试者操作特征曲线(ROC曲线),并计算曲线下面积(AUC),以评估其性能,AUC间比较采用Delong检验。 结果 共纳入609例符合标准的患者,其中114例(18.7%)发生SAP。本研究以苏州大学附属第一医院数据作为训练集(n=426),江苏大学附属金坛医院和苏州永鼎医院数据作为独立测试集(n=183)。多模态模型在测试集中的AUC为0.914,显著高于MCTSI、Ranson评分、BISAP及SABP评分等传统评分系统(AUC分别为0.827、0.675、0.791、0.648),且较深度学习分类模型(AUC=0.685)及影像组学分类模型(AUC=0.739)性能亦有显著提升(基于Delong检验的Z值分别为-3.23、-4.83、-3.48、-4.92、-4.31和-4.59,P值均<0.01)。多模态模型中变量重要性排名前10位的变量依次为胸腔积液、深度学习模型预测值、影像组学模型预测值、甘油三酯、钙离子、SIRS、白细胞计数、年龄、血小板及C反应蛋白,提示上述变量对模型预测SAP具有重要贡献。 结论 本研究基于结构化数据、影像组学特征及深度学习特征,构建了基于XGBoost算法的多中心SAP预测模型,其预测性能优于现有传统评分系统及单模态模型。 -
关键词:
- 胰腺炎, 急性坏死性 /
- 极端梯度提升算法 /
- 影像基因组学 /
- 多模态成像 /
- 深度学习
Abstract:Objective To investigate the application value of a multimodal model integrating radiomics features, deep learning features, and clinical structured data in predicting severe acute pancreatitis (SAP), and to provide more accurate tools for the early identification of SAP in clinical practice. Methods The patients with acute pancreatitis (AP) who attended The First Affiliated Hospital of Soochow University, Jintan Hospital Affiliated to Jiangsu University, and Suzhou Yongding Hospital from January 1, 2017 to December 31, 2023 were included. Related data were collected, including demographic information, previous medical history, etiology, laboratory test data, and systemic inflammatory response syndrome (SIRS) within 24 hours after admission, as well as imaging data within 72 hours after admission, while related scores were calculated, including Ranson score, modified CT severity index (MCTSI), bedside index for severity in acute pancreatitis (BISAP), and systemic inflammatory response syndrome, albumin, blood urea nitrogen and pleural effusion (SABP) score. The model was constructed in the following process: (1) three-dimensional CT images were used to extract and identify radiomics features, and a radiomics classification model was established based on the extreme gradient Boost (XGBoost) algorithm; (2) U-Net is used to perform semantic segmentation of three-dimensional CT images, and then the results of segmentation were imported into 3D ResNet50 to construct a deep learning classification model; (3) the predicted values of the above two models were integrated with clinical structured data to establish a multimodal model based on the XGBoost algorithm. The variable importance plot and local interpretability plot were used to perform visual interpretation of the model. The independent samples t-test was used for comparison of normally distributed continuous data between groups, and the Mann-Whitney U test was used for comparison of non-normally distributed continuous data between groups; the chi-square test or Fisher’s exact test was used for comparison of categorical data between groups. The receiver operating characteristic (ROC) curve was plotted for each model and existing scoring systems, and the area under the ROC curve (AUC) was calculated to assess their performance; the Delong test was used for comparison of AUC. Results A total of 609 patients who met the criteria were included, among whom 114 (18.7%) developed SAP. In this study, the data of 426 patients from The First Affiliated Hospital of Soochow University was used as the training set, and the data of 183 patients from Jintan Hospital Affiliated to Jiangsu University and Suzhou Yongding Hospital were used as the independent test set. The multimodal model had an AUC of 0.914 in the test set, which was significantly higher than the AUC of traditional scoring systems such as MCTSI (AUC=0.827), Ranson score (AUC=0.675), BISAP (AUC=0.791), and SABP score (AUC=0.648); in addition, the multimodal model showed a significant improvement in performance compared with the radiomics classification model (AUC=0.739) and the deep learning classification model (AUC=0.685) (the Delong test: Z=-3.23, -4.83, -3.48, -4.92, -4.31, and -4.59, all P <0.01). The top 10 variables in terms of importance in the multimodal model were pleural effusion, predicted value of the deep learning model, predicted value of the radiomics model, triglycerides, calcium ions, SIRS, white blood cell count, age, platelets, and C-reactive protein, suggesting that the above variables had significant contributions to the performance of the model in predicting SAP. Conclusion Based on structured data, radiomic features, and deep learning features, this study constructs a multicenter prediction model for SAP based on the XGBoost algorithm, which has a better predictive performance than existing traditional scoring systems and unimodal models. -
表 1 纳入研究的患者临床特征
Table 1. The characteristics of the patients in the study
临床特征 训练集 测试集 非SAP组
(n=354)SAP组
(n=72)统计值 P值 非SAP组
(n=141)SAP组
(n=42)统计值 P值 性别[例(%)]
男
277(78.2)
50(69.4)χ2=2.600 0.107
103(73.0)
29(69.0)χ2=0.258 0.258 女 77(21.8) 22(30.6) 38(27.0) 13(31.0) 年龄(岁) 51.51±17.67 50.99±18.18 t=0.225 0.819 52.22±18.04 48.21±17.05 t=1.318 0.203 病因[例(%)]
胆源性
239(67.5)
41(56.9)χ2=2.977 0.395
79(56.0)
22(52.4)0.898 高脂血症性 40(11.3) 11(15.3) 25(17.7) 7(16.7) 酒精性 52(14.7) 14(19.4) 26(18.4) 10(23.8) 其他 23(6.5) 6(8.3) 11(7.8) 3(7.1) 入院CT检查时间(h) 8.05±4.63 7.23±3.19 t=1.434 0.152 8.40±4.77 7.91±3.50 t=0.618 0.537 血小板计数( × 109/L) 198.84±69.03 213.33±75.95 t=-1.498 0.111 202.43±66.39 215.43±80.19 t=-0.957 0.291 白细胞计数( × 109/L) 13.39±6.93 17.23±6.18 t=-4.362 <0.001 13.04±4.79 15.48±6.22 t=-2.343 0.008 钙离子(mmol/L) 2.14±0.16 1.98±0.28 t=4.647 <0.001 2.12±0.15 1.96±0.35 t=3.011 <0.001 甘油三酯(mmol/L) 1.30
(0.83~3.41)2.15
(1.12~6.14)Z=208.606 0.001 1.41
(0.85~3.31)3.26
(1.71~13.78)Z=74.747 <0.001 C反应蛋白(mg/L) 54.85
(8.70~140.93)163.15
(16.22~258.14)Z=189.664 <0.001 49.90
(6.64~144.00)199.39
(22.98~299.10)Z=87.059 <0.001 SIRS[例(%)]
无
248(70.1)
15(20.8)χ2=61.370 <0.001
96(68.1)
8(19.0)χ2=31.726 <0.001 有 106(29.9) 57(79.2) 45(31.9) 34(81.0) 胸腔积液[例(%)]
无
236(66.7)
5(6.9)χ2=86.861 <0.001
95(67.4)
8(19.0)χ2=30.723 <0.001 有 118(33.3) 67(93.1) 46(32.6) 34(81.0) MCTSI(分) 2.00
(2.00~4.00)4.00
(4.00~4.00)Z=117.105 <0.001 2.00
(2.00~4.00)4.00
(4.00~4.00)Z=45.780 <0.001 Ranson评分(分) 1.00
(0.00~2.00)2.00
(1.00~2.00)Z=169.007 <0.001 1.00
(1.00~1.00)2.00
(1.00~2.00)Z=86.654 <0.001 BISAP(分) 1.00
(0.00~2.00)2.00
(2.00~3.00)Z=88.789 <0.001 1.00
(0.00~2.00)2.00
(2.00~3.00)Z=55.616 <0.001 SABP评分(分) 2.70
(-3.09~8.92)10.09
(3.01~25.77)Z=176.081 <0.001 3.89
(-2.80~11.76)9.00
(1.15~21.31)Z=93.856 0.004 表 2 各个模型与已有评分系统在数据集上的表现
Table 2. Performance of the models and existing scoring systems in datasets
分类 指标 MCTSI Ranson评分 BISAP SABP评分 深度学习 影像组学 多模态模型 训练集 AUC 0.786 0.694 0.840 0.682 0.655 0.692 0.926 P值 <0.01 <0.01 <0.01 <0.01 <0.01 <0.01 Z值 -6.11 -6.49 -4.32 -5.76 -7.22 -7.36 敏感度 0.861 0.583 0.875 0.583 0.514 0.792 0.847 特异度 0.678 0.743 0.706 0.715 0.850 0.599 0.884 测试集 AUC 0.827 0.675 0.791 0.648 0.685 0.739 0.914 P值 <0.01 <0.01 <0.01 <0.01 <0.01 <0.01 Z值 -3.23 -4.83 -3.48 -4.92 -4.31 -4.59 敏感度 0.929 0.524 0.762 0.357 0.571 0.881 0.905 特异度 0.679 0.751 0.716 0.879 0.844 0.560 0.823 注:P值和Z值是多模态模型与其他评分或模型AUC的Delong检验结果。
-
[1] PETROV MS, YADAV D. Global epidemiology and holistic prevention of pancreatitis[J]. Nat Rev Gastroenterol Hepatol, 2019, 16( 3): 175- 184. DOI: 10.1038/s41575-018-0087-5. [2] TRIKUDANATHAN G, YAZICI C, EVANS PHILLIPS A, et al. Diagnosis and management of acute pancreatitis[J]. Gastroenterology, 2024, 167( 4): 673- 688. DOI: 10.1053/j.gastro.2024.02.052. [3] MEDEROS MA, REBER HA, GIRGIS MD. Acute pancreatitis[J]. Jama, 2021, 325( 4): 382. DOI: 10.1001/jama.2020.20317. [4] YIN MY, ZHU JD, LIU L, et al. Research advances in machine learning models for acute pancreatitis[J]. J Clin Hepatol, 2023, 39( 12): 2978- 2984. DOI: 10.3969/j.issn.1001-5256.2023.12.034.殷民月, 朱锦舟, 刘璐, 等. 急性胰腺炎机器学习模型的研究进展[J]. 临床肝胆病杂志, 2023, 39( 12): 2978- 2984. DOI: 10.3969/j.issn.1001-5256.2023.12.034. [5] XIE F, ZHANG KQ, LI F, et al. Diagnostic accuracy of convolutional neural network-based endoscopic image analysis in diagnosing gastric cancer and predicting its invasion depth: A systematic review and meta-analysis[J]. Gastrointest Endosc, 2022, 95( 4): 599- 609. DOI: 10.1016/j.gie.2021.12.021. [6] HAGGENMÜLLER S, MARON RC, HEKLER A, et al. Skin cancer classification via convolutional neural networks: Systematic review of studies involving human experts[J]. Eur J Cancer, 2021, 156: 202- 216. DOI: 10.1016/j.ejca.2021.06.049. [7] LI GW, LIU J, CAO H, et al. Research review of deep learning in colon polyp image segmentation[J]. J Front Comput Sci Technoly, 2025, 19( 5): 1198- 1216. DOI: 10.3778/j.issn.1673-9418.2408012.李国威, 刘静, 曹慧, 等. 深度学习在结肠息肉图像分割中的研究综述[J]. 计算机科学与探索, 2025, 19( 5): 1198- 1216. DOI: 10.3778/j.issn.1673-9418.2408012. [8] PACAL I, KARABOGA D. A robust real-time deep learning based automatic polyp detection system[J]. Comput Biol Med, 2021, 134: 104519. DOI: 10.1016/j.compbiomed.2021.104519. [9] YU T, LIN N, ZHANG X, et al. An end-to-end tracking method for polyp detectors in colonoscopy videos[J]. Artif Intell Med, 2022, 131: 102363. DOI: 10.1016/j.artmed.2022.102363. [10] LIU H, ZHUANG YZ, SONG EM, et al. A 3D boundary-guided hybrid network with convolutions and Transformers for lung tumor segmentation in CT images[J]. Comput Biol Med, 2024, 180: 109009. DOI: 10.1016/j.compbiomed.2024.109009. [11] BANKS PA, BOLLEN TL, DERVENIS C, et al. Classification of acute pancreatitis: 2012: Revision of the Atlanta classification and definitions by international consensus[J]. Gut, 2013, 62( 1): 102- 111. DOI: 10.1136/gutjnl-2012-302779. [12] KLINE A, WANG HY, LI YK, et al. Multimodal machine learning in precision health: A scoping review[J]. NPJ Digit Med, 2022, 5( 1): 171. DOI: 10.1038/s41746-022-00712-8. [13] LIPKOVA J, CHEN RJ, CHEN BW, et al. Artificial intelligence for multimodal data integration in oncology[J]. Cancer Cell, 2022, 40( 10): 1095- 1110. DOI: 10.1016/j.ccell.2022.09.012. [14] MOHSEN F, ALI H, HAJJ N EL, et al. Artificial intelligence-based methods for fusion of electronic health records and imaging data[J]. Sci Rep, 2022, 12( 1): 17981. DOI: 10.1038/s41598-022-22514-4. [15] SHAO J, MA JC, ZHANG Q, et al. Predicting gene mutation status via artificial intelligence technologies based on multimodal integration(MMI) to advance precision oncology[J]. Semin Cancer Biol, 2023, 91: 1- 15. DOI: 10.1016/j.semcancer.2023.02.006. [16] HUANG SC, PAREEK A, SEYYEDI S, et al. Fusion of medical imaging and electronic health records using deep learning: A systematic review and implementation guidelines[J]. NPJ Digit Med, 2020, 3: 136. DOI: 10.1038/s41746-020-00341-z. [17] GLIEM N, AMMER-HERRMENAU C, ELLENRIEDER V, et al. Management of severe acute pancreatitis: An update[J]. Digestion, 2021, 102( 4): 503- 507. DOI: 10.1159/000506830. [18] GAO X, LIN JX, WU AR, et al. Application of machine learning model based on XGBoost algorithm in early prediction of patients with acute severe pancreatitis[J]. Chin Crit Care Med, 2023, 35( 4): 421- 426. DOI: 10.3760/cma.j.cn121430-20221019-00930.高欣, 林嘉希, 吴爱荣, 等. 基于XGBoost算法的机器学习模型在早期预测重症急性胰腺炎中的应用[J]. 中华危重病急救医学, 2023, 35( 4): 421- 426. DOI: 10.3760/cma.j.cn121430-20221019-00930. [19] CHEN ZY, WANG Y, ZHANG HL, et al. Deep learning models for severity prediction of acute pancreatitis in the early phase from abdominal nonenhanced computed tomography images[J]. Pancreas, 2023, 52( 1): e45- e53. DOI: 10.1097/MPA.0000000000002216. [20] YIN MY, LIN JX, WANG Y, et al. Development and validation of a multimodal model in predicting severe acute pancreatitis based on radiomics and deep learning[J]. Int J Med Inform, 2024, 184: 105341. DOI: 10.1016/j.ijmedinf.2024.105341. [21] KAMNITSAS K, LEDIG C, NEWCOMBE VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation[J]. Med Image Anal, 2017, 36: 61- 78. DOI: 10.1016/j.media.2016.10.004. [22] LITJENS G, KOOI T, BEJNORDI BE, et al. A survey on deep learning in medical image analysis[J]. Med Image Anal, 2017, 42: 60- 88. DOI: 10.1016/j.media.2017.07.005. -



 PDF下载 ( 1541 KB)
PDF下载 ( 1541 KB)


 下载:
下载: