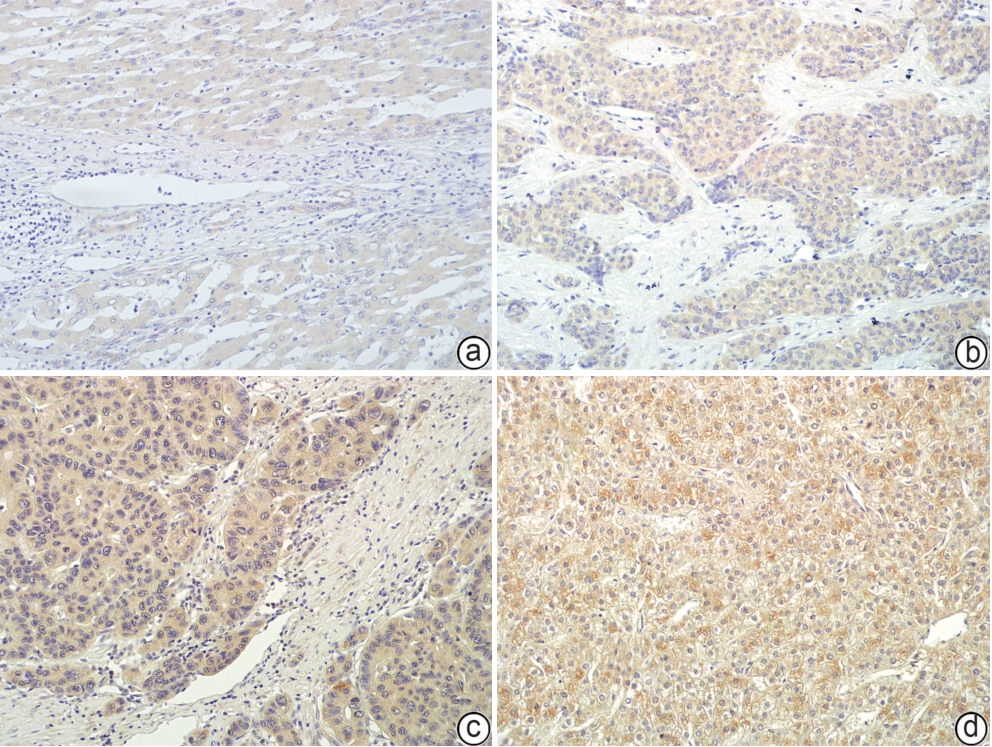
C1q肿瘤坏死因子相关蛋白3(C1QTNF3)在肝癌中的表达及其对预后的预测价值
DOI: 10.12449/JCH250520
-
摘要:
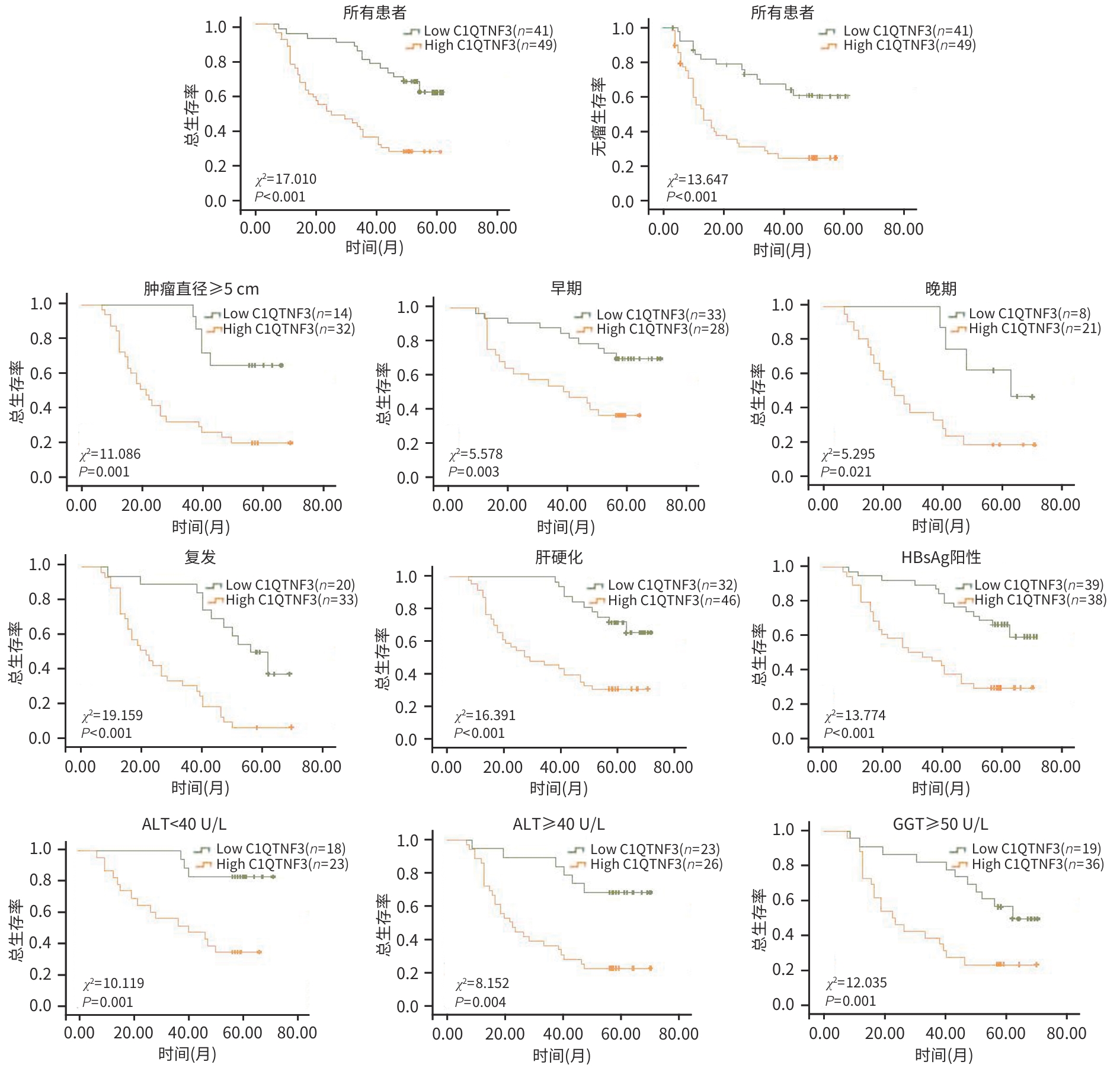
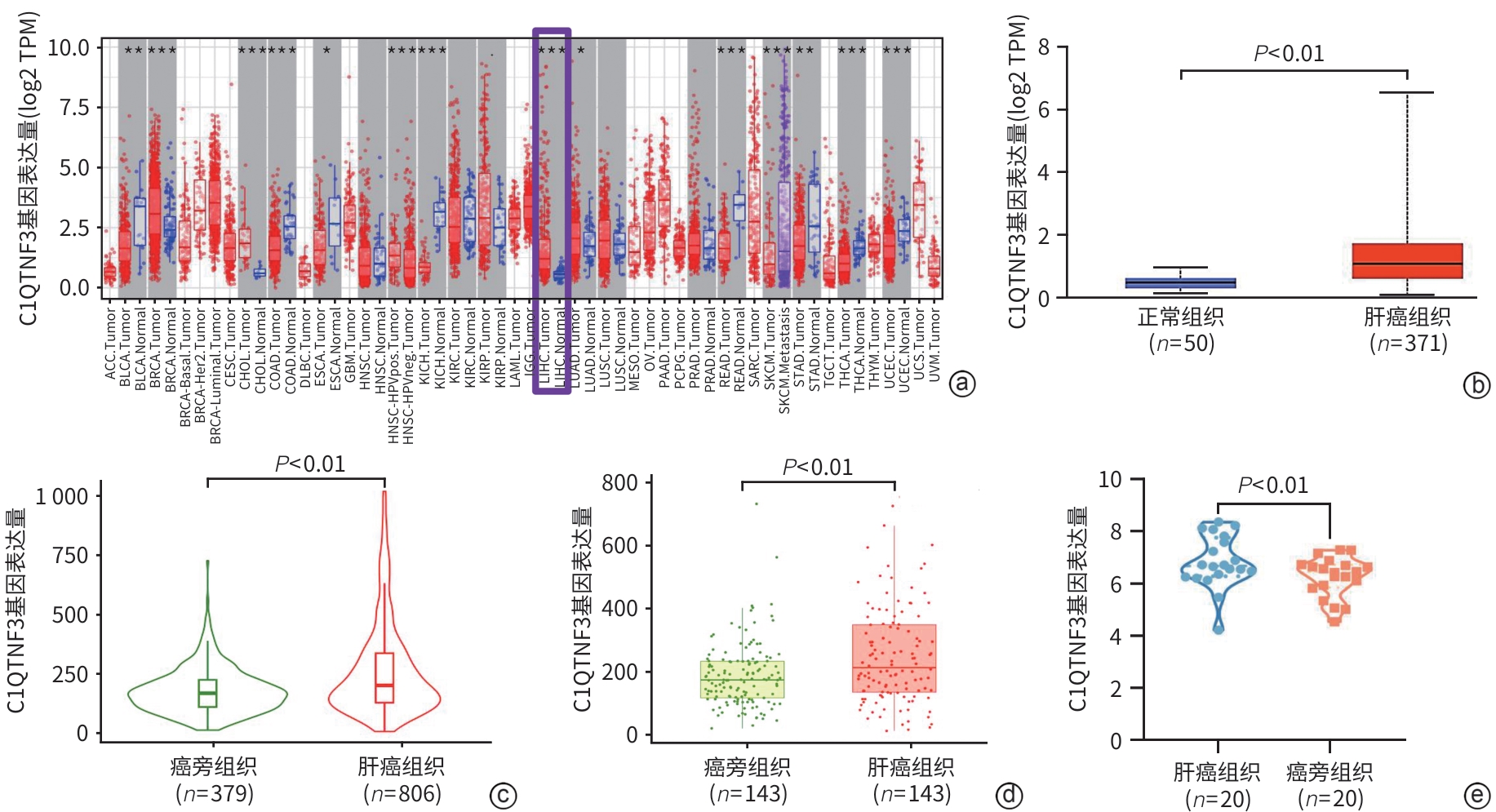
目的 探究C1q肿瘤坏死因子相关蛋白3(C1QTNF3)在肝癌组织中的表达情况,分析其与患者临床病理特征的关系,并进一步评估其在肝癌预后中的潜在预测价值。 方法 分别于TIMER、UALCAN、TNMplot及GEO数据库收集资料,采用生物信息学分析C1QTNF3基因在泛癌、正常组织与肝癌组织,以及癌组织及其癌旁组织中的表达水平。选取90例肝癌患者的肝癌组织及其癌旁组织标本,收集患者年龄、性别、肿瘤直径、肿瘤数目等临床资料。计量资料组间比较采用成组t检验或配对t检验;计数资料组间比较采用χ2检验。Kaplan-Meier法绘制生存曲线,Log-rank检验分析C1QTNF3表达水平与肝癌患者生存期的关系。Cox回归模型分析影响肝癌患者预后的危险因素,受试者操作特征曲线(ROC曲线)分析不同时间点C1QTNF3表达对肝癌患者预后的预测能力。 结果 生物信息学分析结果显示,C1QTNF3基因在多种恶性肿瘤组织中表达上调,尤其在肝癌组织中(P<0.001);C1QTNF3基因在肝癌组织中的表达量高于正常组织及其癌旁组织(P值均<0.01)。90例肝癌患者免疫组化染色结果显示,C1QTNF3主要表达于细胞质,少量表达于细胞核,且在癌旁组织中主要呈阴性表达,在肝癌组织中呈阳性表达。C1QTNF3蛋白在肝癌组织中的阳性表达率(76.67% vs 33.33%)和强阳性表达率(54.44% vs 5.56%)均显著高于癌旁组织(χ2值分别为34.141、51.217,P值均<0.01)。肿瘤直径≥5 cm、晚期、肝硬化、HBsAg阴性和GGT≥50 U/L肝癌患者的C1QTNF3蛋白强阳性表达率均显著高于肿瘤直径<5 cm、早期、无肝硬化、HBsAg阳性和GGT<50 U/L的肝癌患者(P值均<0.05)。单因素Cox回归分析结果显示,肿瘤直径、复发情况和C1QTNF3表达是肝癌患者预后的影响因素(P值均<0.05);多因素Cox回归分析结果显示,C1QTNF3表达水平和复发情况是影响肝癌患者生存期的独立危险因素(P值均<0.05)。生存曲线结果显示,所有肝癌患者中,C1QTNF3高表达(强阳性)者相较于低表达者,总生存率和无瘤生存率均更短(χ2值分别为17.010、13.647,P值均<0.001);在肿瘤直径≥5 cm、早/晚期、复发、肝硬化、HBsAg阳性、ALT<40 U/L、ALT≥40 U/L和GGT≥50 U/L的肝癌患者中,C1QTNF3高表达者的总生存率均显著降低(χ2值分别11.086、5.578、5.295、19.159、16.391、13.774、10.119、8.152、12.035,P值均<0.05)。ROC曲线结果显示,C1QTNF3表达在5年时的预测潜力最强,ROC曲线下面积为0.77。 结论 C1QTNF3在肝癌组织中呈高表达,其表达水平和复发情况与肝癌患者的生存期密切相关,C1QTNF3蛋白高表达患者生存率更低。 Abstract:Objective To investigate the expression of C1q tumor necrosis factor-related protein 3 (C1QTNF3) in liver cancer tissue, its association with the clinicopathological features of patients, and its potential value in predicting the prognosis of liver cancer. Methods Related data were collected from TIMER, UALCAN, TNMplot, and GEO databases, and the bioinformatics methods were used to measure the expression level of C1QTNF3 in pan-cancer, normal tissue/liver cancer tissue, and cancerous tissue/paracancerous tissue. Cancerous and paracancerous tissue samples were collected from 90 patients with liver cancer, and related clinical data were collected, including age, sex, tumor diameter, and tumor number. The independent-samples t test or the paired t-test was used for comparison of continuous data between groups, and the chi-square test was used for comparison of categorical data between groups. The Kaplan-Meier method was used to plot survival curves, and the Log-rank test was used to investigate the association between the expression level of C1QTNF3 and the survival of patients with liver cancer. The Cox regression model was used to identify the risk factors for the prognosis of patients with liver cancer, and the receiver operating characteristic (ROC) curve was used to analyze the ability of C1QTNF3 expression at different time points for predicting the prognosis of patients with liver cancer. Results The bioinformatics analysis showed that the expression of C1QTNF3 was upregulated in various malignant tumors, especially in liver cancer tissue (P<0.001), and the expression level of C1QTNF3 in liver cancer tissue was significantly higher than that in normal tissue and paracancerous tissues (all P<0.01). The immunohistochemical staining results of 90 patients with liver cancer showed that C1QTNF3 was mainly expressed in cytoplasm, with a small amount in nucleus, and it had negative expression in paracancerous tissue and positive expression in liver cancer tissue. The positive expression rate and strong positive expression rate of C1QTNF3 protein in liver cancer tissue were significantly higher than those in paracancerous tissue (positive expression rate: 76.67% vs 33.33%, χ2=34.141, P<0.01; strong positive expression rate: 54.44% vs 5.56%, χ2=51.217, P<0.01). The liver cancer patients with a tumor diameter of ≥5 cm, an advanced stage, the presence of liver cirrhosis, negative HBsAg, and gamma-glutamyl transpeptidase (GGT)≥50 U/L had a significantly higher strong positive expression rate of C1QTNF3 protein than those with a tumor diameter of <5 cm, an early stage, the absence of liver cirrhosis, positive HBsAg, and GGT<50 U/L (all P<0.05). The univariate Cox regression analysis showed that tumor diameter, recurrence, and C1QTNF3 expression were influencing factors for the prognosis of patients with liver cancer (all P<0.05), and the multivariate Cox regression analysis showed that the expression level of C1QTNF3 and recurrence were independent risk factors for the survival of patients with liver cancer (both P<0.05). The survival curve analysis showed that for all patients with liver cancer, the patients with high (strong positive) expression of C1QTNF3 had significantly lower overall survival rate and disease-free survival rate than those with low expression (χ2=17.010 and 13.647, both P<0.001); for liver cancer patients with a tumor diameter of ≥5 cm, an early/advanced stage, recurrence, the presence of liver cirrhosis, positive HBsAg, alanine aminotransferase (ALT) <40 U/L, ALT≥40 U/L, and GGT≥50 U/L, the patients with high expression of C1QTNF3 had a significant reduction in overall survival rate (χ2=11.086, 5.578, 5.295, 19.159, 16.391, 13.774, 10.119, 8.152, and 12.035, all P<0.05). The ROC curve analysis showed that C1QTNF3 expression had the strongest predictive potential at 5 years, with an area under the ROC curve of 0.77. Conclusion C1QTNF3 is highly expressed in liver cancer tissue, and the expression level of C1QTNF3 and recurrence are closely associated with the survival of patients with liver cancer. Patients with high expression of C1QTNF3 protein tend to have a lower survival rate. -
Key words:
- Liver Neoplasms /
- Adipokines /
- Prognosis
-
表 1 C1QTNF3在肝癌组织及癌旁组织中阳性表达率和强阳性表达率的比较
Table 1. Comparison of the positive expression rate and the strong positive expression rate of C1QTNF3 in liver cancer tissues and adjacent tissues
C1QTNF3蛋白 肝癌组织 (n=90) 癌旁组织 (n=90) χ2值 P值 阳性率[例(%)] 69(76.67) 30(33.33) 34.141 <0.01 强阳性率[例(%)] 49(54.44) 5(5.56) 51.217 <0.01 表 2 C1QTNF3蛋白高表达与肝癌临床病理特征之间的关系
Table 2. The relationship between high expression of C1QTNF3 protein and clinicopathological features of liver cancer
临床病理特征 例数 强阳性[例(%)] χ2值 P值 性别 0.155 0.694 男 74 41(55.4) 女 16 8(50.0) 年龄 0.278 0.598 ≥50岁 39 20(51.3) <50岁 51 29(56.9) 肿瘤直径 8.674 0.003 <5 cm 44 17(38.6) ≥5 cm 46 32(69.6) 肿瘤数目 0.026 0.873 ≤1个 74 40(54.1) >1个 16 9(56.3) 包膜 0.453 0.501 有 47 24(51.1) 无 43 25(58.1) 组织学分级 0.487 0.485 Ⅰ~Ⅱ级 58 30(51.7) Ⅲ级 32 19(59.4) 临床分期 5.570 0.018 早期 61 28(45.9) 晚期 29 21(72.4) 复发情况 3.178 0.075 有 53 33(62.3) 无 37 16(43.2) 肝硬化 4.840 0.028 有 78 46(59.0) 无 12 3(25.0) HBsAg 5.979 0.014 阳性 77 38(49.4) 阴性 13 11(84.6) 抗-HBc 2.962 0.085 阳性 80 41(51.3) 阴性 10 8(80.0) 抗-HCV 0.016 0.898 阳性 2 1(50.0) 阴性 88 48(54.5) TBil 3.789 0.052 <17.1 μmol/L 66 40(60.6) ≥17.1 μmol/L 24 9(37.5) ALT 0.083 0.773 <40 U/L 41 23(56.1) ≥40 U/L 49 26(53.1) AFP 0.000 0.988 <400 μg/L 57 31(54.4) ≥400 μg/L 33 18(54.5) GGT 6.912 0.009 <50 U/L 35 13(37.1) ≥50 U/L 55 36(65.5) 表 3 肝癌患者预后Cox风险回归模型单因素分析
Table 3. Univariate analysis of Cox risk regression model for prognosis of liver cancer patients
参数 B值 SE Wald HR(95%CI) P值 性别 0.384 0.584 1.828 1.468(0.841~2.561) 0.176 年龄 0.042 0.215 0.038 1.043(0.684~1.589) 0.846 肿瘤直径 0.548 0.214 6.541 1.730(1.137~2.634) 0.011 分化程度 0.023 0.222 0.011 0.977(0.633~1.510) 0.918 临床分期 0.223 0.229 0.949 0.800(0.511~1.253) 0.330 肿瘤数目 0.110 0.278 0.156 0.896(0.520~1.544) 0.692 包膜 0.002 0.213 0.000 0.998(0.658~1.515) 0.993 复发情况 0.740 0.221 11.206 2.095(1.359~3.230) 0.001 肝硬化 0.378 0.316 1.431 0.686(0.369~1.273) 0.232 抗-HBc 0.142 0.339 0.177 0.867(0.447~1.684) 0.674 抗-HCV 0.603 0.724 0.695 0.547(0.132~2.260) 0.404 TBil 0.036 0.240 0.022 0.965(0.603~1.543) 0.881 ALT 0.021 0.215 0.010 1.022(0.670~1.558) 0.921 AFP 0.378 0.221 2.911 1.459(0.945~2.252) 0.088 GGT 0.313 0.219 2.038 1.367(0.890~2.101) 0.153 HBsAg 0.252 0.303 0.691 0.777(0.430~1.407) 0.406 C1QTNF3表达 0.788 0.218 13.038 2.198(1.434~3.371) <0.001 表 4 肝癌患者预后Cox风险回归模型多因素分析
Table 4. Multivariate analysis of Cox risk regression model for prognosis of liver cancer patients
参数 B值 SE Wald HR(95%CI) P值 ALT 0.292 0.240 1.482 0.747(0.467~1.195) 0.223 肿瘤直径 0.274 0.232 1.399 1.315(0.835~2.072) 0.237 复发情况 0.835 0.249 11.249 2.305(1.415~3.756) 0.001 C1QTNF3表达 0.701 0.233 9.060 2.016(1.277~3.182) 0.003 -
[1] CHEN WQ, ZHENG RS, BAADE PD, et al. Cancer statistics in China, 2015[J]. CA Cancer J Clin, 2016, 66( 2): 115- 132. DOI: 10.3322/caac.21338. [2] HOU ZY, LIU Y, YANG CR, et al. Current research status and prospect of circulating tumor DNA in hepatocellular carcinoma[J]. J Clin Hepatol, 2022, 38( 11): 2616- 2620. DOI: 10.3969/j.issn.1001-5256.2022.11.036.侯志远, 刘源, 杨超然, 等. 循环肿瘤DNA在肝细胞癌中的研究现状及展望[J]. 临床肝胆病杂志, 2022, 38( 11): 2616- 2620. DOI: 10.3969/j.issn.1001-5256.2022.11.036. [3] RAN XK, CHEN XJ, ZHAO YX, et al. Expression and clinical significance of forkhead box A2 and forkhead box J2 in hepatocellular carcinoma[J]. J Clin Hepatol, 2021, 37( 6): 1342- 1347. DOI: 10.3969/j.issn.1001-5256.2021.06.025.冉小柯, 陈欣菊, 赵云霞, 等. 叉头转录因子A2、J2在肝细胞癌组织中的表达及意义[J]. 临床肝胆病杂志, 2021, 37( 6): 1342- 1347. DOI: 10.3969/j.issn.1001-5256.2021.06.025. [4] YU XH, CHEN BD, LIU YM, et al. Role of NOD-like receptor protein 3 inflammasome in the development and progression of hepatocellular carcinoma[J]. J Clin Hepatol, 2024, 40( 2): 397- 401. DOI: 10.12449/JCH240229.余学海, 陈本栋, 刘伊敏, 等. NOD样受体蛋白3(NLRP3)炎性小体在肝细胞癌发生发展中的作用[J]. 临床肝胆病杂志, 2024, 40( 2): 397- 401. DOI: 10.12449/JCH240229. [5] LI JJ, YANG HH, HUO G. Analysis of clinical features, cell morphology and prognostic factors in patients with primary liver cancer[J]. J Clin Exp Med, 2024, 23( 6): 566- 570. DOI: 10.3969/j.issn.1671-4695.2024.06.002.李姣姣, 杨会会, 霍刚. 原发性肝癌患者临床特征、细胞形态学分析及其预后的影响因素分析[J]. 临床和实验医学杂志, 2024, 23( 6): 566- 570. DOI: 10.3969/j.issn.1671-4695.2024.06.002. [6] GUO P, LU W, YANG WX, et al. Research progress in biomarkers of drug-induced liver injury[J]. J Clin Hepatol, 2016, 32( 9): 1822- 1826. DOI: 10.3969/j.issn.1001-5256.2016.09.043.郭佩, 卢旺, 杨文轩, 等. 药物性肝损伤相关生物学标志物的研究进展[J]. 临床肝胆病杂志, 2016, 32( 9): 1822- 1826. DOI: 10.3969/j.issn.1001-5256.2016.09.043. [7] ALBA MM, EBRIGHT B, HUA B, et al. Eicosanoids and other oxylipins in liver injury, inflammation and liver cancer development[J]. Front Physiol, 2023, 14: 1098467. DOI: 10.3389/fphys.2023.1098467. [8] NATI M, CHUNG KJ, CHAVAKIS T. The role of innate immune cells in nonalcoholic fatty liver disease[J]. J Innate Immun, 2022, 14( 1): 31- 41. DOI: 10.1159/000518407. [9] YAN RJ, JIAO JZ, HUANG Y, et al. Research advances in liver macrophages regulating malignant transformation of hepatic precancerous lesions[J]. J Clin Hepatol, 2024, 40( 5): 1039- 1043. DOI: 10.12449/JCH240527.闫瑞娟, 焦俊喆, 黄玉, 等. 肝巨噬细胞调控肝癌癌前病变恶变的研究进展[J]. 临床肝胆病杂志, 2024, 40( 5): 1039- 1043. DOI: 10.12449/JCH240527. [10] KHURANA A, NAVIK U, ALLAWADHI P, et al. Spotlight on liver macrophages for halting liver disease progression and injury[J]. Expert Opin Ther Targets, 2022, 26( 8): 707- 719. DOI: 10.1080/14728222.2022.2133699. [11] MAO ZF, YANG LH, LU XS, et al. C1QTNF3 in the murine ovary and its function in folliculogenesis[J]. Reproduction, 2018, 155( 4): 333- 346. DOI: 10.1530/REP-17-0783. [12] MICALLEF P, VUJIČIĆ M, WU Y, et al. C1QTNF3 is upregulated during subcutaneous adipose tissue remodeling and stimulates macrophage chemotaxis and M1-like polarization[J]. Front Immunol, 2022, 13: 914956. DOI: 10.3389/fimmu.2022.914956. [13] ZHENG WP, SHEN ZY. Risk factors for tumor recurrence after liver transplantation for hepatocellular carcinoma and related control strategies[J]. J Clin Hepatol, 2019, 35( 11): 2391- 2395. DOI: 10.3969/j.issn.1001-5256.2019.11.003.郑卫萍, 沈中阳. 肝细胞癌肝移植术后肿瘤复发的危险因素与防治策略[J]. 临床肝胆病杂志, 2019, 35( 11): 2391- 2395. DOI: 10.3969/j.issn.1001-5256.2019.11.003. [14] WANG TR, XU AG, CHEN L, et al. Values of united determining serum gangliosides and fetoprotein-A on primary hepatic carcinoma diagnosis[J]. J Clin Hepatol, 2006, 22( 4): 254- 255. DOI: 10.3969/j.issn.1001-5256.2006.04.005.王天然, 许爱国, 陈莉, 等. 血清GLS和AFP联合测定对原发性肝癌诊断的意义[J]. 临床肝胆病杂志, 2006, 22( 4): 254- 255. DOI: 10.3969/j.issn.1001-5256.2006.04.005. [15] YANG GM, ZHAO YS, WANG CH, et al. Research advances in tumor markers for the diagnosis of hepatocellular carcinoma[J]. J Clin Hepatol, 2018, 34( 1): 199- 203. DOI: 10.3969/j.issn.1001-5256.2018.01.044.杨贵敏, 赵运胜, 王春华, 等. 肝细胞癌肿瘤标志物的研究进展[J]. 临床肝胆病杂志, 2018, 34( 1): 199- 203. DOI: 10.3969/j.issn.1001-5256.2018.01.044. [16] ZHU MY, CHEN J, ZHANG XX. Research advances in serum biomarkers for early diagnosis of hepatocellular carcinoma[J]. J Clin Hepatol, 2014, 30( 10): 1091- 1093. DOI: 10.3969/j.issn.1001-5256.2014.10.029.朱明玉, 陈洁, 张欣欣. 肝细胞癌早期诊断血清学标志物的研究进展[J]. 临床肝胆病杂志, 2014, 30( 10): 1091- 1093. DOI: 10.3969/j.issn.1001-5256.2014.10.029. [17] YIN JY, WANG Q. Progress on adipokines in non-alcoholic fatty liver disease[J/CD]. Chin J Liver Dis(Electronic Version), 2023, 15( 1): 1- 5. DOI: 10.3969/j.issn.1674-7380.2023.01.001.尹静亚, 王琦. 脂肪因子在非酒精性脂肪性肝病中研究进展[J/CD]. 中国肝脏病杂志(电子版), 2023, 15( 1): 1- 5. DOI: 10.3969/j.issn.1674-7380.2023.01.001. [18] SCHERER PE, WILLIAMS S, FOGLIANO M, et al. A novel serum protein similar to C1q, produced exclusively in adipocytes[J]. J Biol Chem, 1995, 270( 45): 26746- 26749. DOI: 10.1074/jbc.270.45.26746. [19] WEI SC, ZHENG GQ. Role of adipokine in the pathogenesis of cirrhosis of the liver[J]. J Clin Hepatol, 2012, 28( 1): 78- 81.魏思忱, 郑国启. 脂肪因子在肝硬化发病机制中的作用[J]. 临床肝胆病杂志, 2012, 28( 1): 78- 81. [20] SHAPIRO L, SCHERER PE. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor[J]. Curr Biol, 1998, 8( 6): 335- 338. DOI: 10.1016/s0960-9822(98)70133-2. [21] BYERLY MS, PETERSEN PS, RAMAMURTHY S, et al. C1q/TNF-related protein 4(CTRP4) is a unique secreted protein with two tandem C1q domains that functions in the hypothalamus to modulate food intake and body weight[J]. J Biol Chem, 2014, 289( 7): 4055- 4069. DOI: 10.1074/jbc.M113.506956. [22] BYERLY MS, SWANSON R, WEI ZK, et al. A central role for C1q/TNF-related protein 13(CTRP13) in modulating food intake and body weight[J]. PLoS One, 2013, 8( 4): e62862. DOI: 10.1371/journal.pone.0062862. [23] KAMBARA T, OHASHI K, SHIBATA R, et al. CTRP9 protein protects against myocardial injury following ischemia-reperfusion through AMP-activated protein kinase(AMPK)-dependent mechanism[J]. J Biol Chem, 2012, 287( 23): 18965- 18973. DOI: 10.1074/jbc.M112.357939. [24] KLONISCH T, GLOGOWSKA A, THANASUPAWAT T, et al. Structural commonality of C1q TNF-related proteins and their potential to activate relaxin/insulin-like family peptide receptor 1 signalling pathways in cancer cells[J]. Br J Pharmacol, 2017, 174( 10): 1025- 1033. DOI: 10.1111/bph.13559. [25] ELSAID HH, ELGOHARY MN, ELSHABRAWY AM. Complement c1q tumor necrosis factor-related protein 3 a novel adipokine, protect against diabetes mellitus in young adult Egyptians[J]. Diabetes Metab Syndr, 2019, 13( 1): 434- 438. DOI: 10.1016/j.dsx.2018.10.004. [26] CANDLER TP, MAHMOUD O, LYNN RM, et al. Continuing rise of type 2 diabetes incidence in children and young people in the UK[J]. Diabet Med, 2018, 35( 6): 737- 744. DOI: 10.1111/dme.13609. [27] YARIBEYGI H, ATKIN SL, SAHEBKAR A. Mitochondrial dysfunction in diabetes and the regulatory roles of antidiabetic agents on the mitochondrial function[J]. J Cell Physiol, 2019, 234( 6): 8402- 8410. DOI: 10.1002/jcp.27754. [28] YARIBEYGI H, RASHIDFARROKHI F, ATKIN SL, et al. C1q/TNF-related protein-3 and glucose homeostasis[J]. Diabetes Metab Syndr, 2019, 13( 3): 1923- 1927. DOI: 10.1016/j.dsx.2019.04.047. [29] WILLIAM WONG G, KRAWCZYK SA, KITIDIS-MITROKOSTAS C, et al. Molecular, biochemical and functional characterizations of C1q/TNF family members: Adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions[J]. Biochem J, 2008, 416( 2): 161- 177. DOI: 10.1042/BJ20081240. [30] MAEDA T, ABE M, KURISU K, et al. Molecular cloning and characterization of a novel gene, CORS26 encoding a putative secretory protein and its possible involvement in skeletal development[J]. J Biol Chem, 2001, 276( 5): 3628- 3634. DOI: 10.1074/jbc.M007898200. [31] CHEN Q, LAI SM, XU SH, et al. Resident macrophages restrain pathological adipose tissue remodeling and protect vascular integrity in obese mice[J]. EMBO Rep, 2021, 22( 8): e52835. DOI: 10.15252/embr.202152835. [32] DIRAT B, BOCHET L, DABEK M, et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion[J]. Cancer Res, 2011, 71( 7): 2455- 2465. DOI: 10.1158/0008-5472.CAN-10-3323. [33] NEVOLA R, RUOCCO R, CRISCUOLO L, et al. Predictors of early and late hepatocellular carcinoma recurrence[J]. World J Gastroenterol, 2023, 29( 8): 1243- 1260. DOI: 10.3748/wjg.v29.i8.1243. [34] YANG ZW, SONG YX, CAO CY, et al. Research progress on effects of C1q-positive tumor-associated macrophages on T cell metabolism and function[J]. China Ind Econ, 2024, 30( 11): 907- 916. DOI: 10.11735/j.issn.1671-170X.2024.11.B004.杨子薇, 宋羽霄, 曹春雨, 等. C1q阳性肿瘤相关巨噬细胞影响T细胞代谢及功能的研究进展[J]. 肿瘤学杂志, 2024, 30( 11): 907- 916. DOI: 10.11735/j.issn.1671-170X.2024.11.B004. -



 PDF下载 ( 4068 KB)
PDF下载 ( 4068 KB)

 下载:
下载: