非酒精性脂肪性肝病多组学研究现状
DOI: 10.12449/JCH240626
-
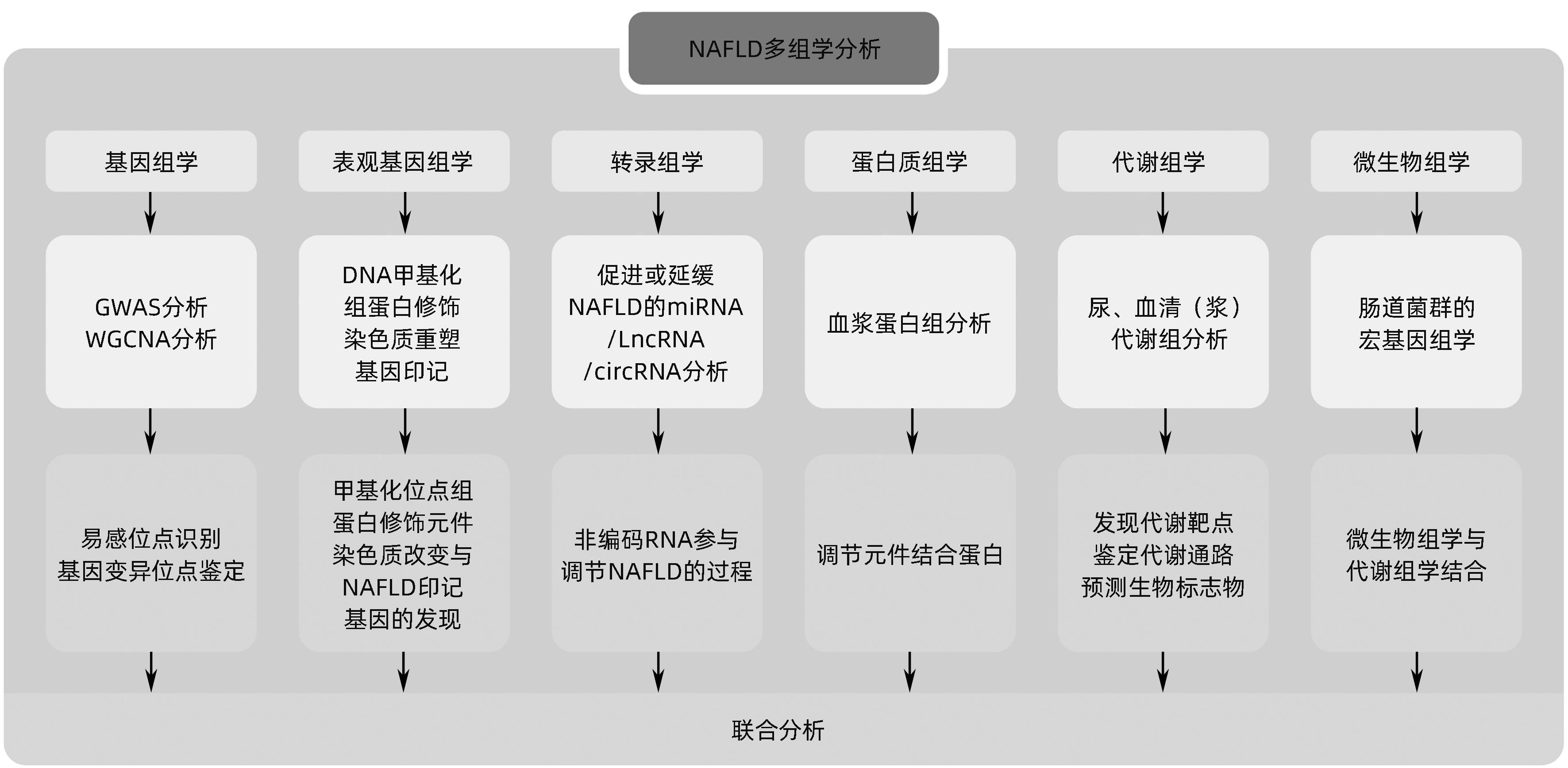
摘要: 非酒精性脂肪性肝病(NAFLD)在全球范围内患病率高达30%,严重影响人类健康并构成公共卫生负担。由于该病难以诊断和监控,因此,识别潜在的药物靶点和生物标志物具有重要价值,多组学技术在探索NAFLD早期诊断标志物、治疗靶点、疗效和预后评估方面具有广阔的前景。本文对近年来多组学技术在NAFLD中领域的研究进展进行综述,以期为NAFLD的防治提供更为丰富的理论依据和新的策略。Abstract: The prevalence rate of nonalcoholic fatty liver disease (NAFLD) reaches up to 30% around the world, and the disease has a serious impact on human health and constitutes a public health burden. Due to difficulties in the diagnosis and monitoring of NAFLD, it is important to identify potential drug targets and biomarkers, and multi-omics techniques hold great promise in the search for early diagnostic markers, therapeutic targets, and outcome and prognostic assessment of NAFLD. This article reviews the research advances in multi-omics techniques in the field of NAFLD in recent years, in order to provide a richer theoretical basis and new strategies for the prevention and treatment of NAFLD.
-
Key words:
- Non-alcoholic Fatty Liver Disease /
- Genomics /
- Proteomics /
- Metabolomics /
- Gastrointestinal Microbiome
-
表 1 NAFLD的代谢组学研究
Table 1. Metabolomics studies in NAFLD
样本类型 组别 重要代谢物 涉及代谢通路 参考文献 血清 NAFLD组(n=157),NASH组(n=138),健康对照组(n=66) 脂肪酸;甘油酯;甘油磷脂,溶血磷脂酸、溶血磷脂酰胆碱、溶血磷脂酰乙醇胺、磷脂酰胆碱、磷脂酰乙醇胺和磷脂酰肌醇;鞘脂 脂质代谢 [53] 血清 NAFLD组(n=144),健康对照组(n=368) s-腺苷蛋氨酸、s-腺苷同型半胱氨酸和同型半胱氨酸 氨基酸代谢、脂质代谢 [56] 血浆 NAFLD组(n=132),健康对照组(n=42) 磷脂酰胆碱、溶血磷脂酰乙醇胺、磷脂酰胆碱、天冬氨酸转氨酶、丙氨酸转氨酶、γ-谷氨酰转肽酶、白蛋白、总胆红素、甘油三酯、总胆固醇、低密度脂蛋白、胆碱酯酶、透明质酸、C反应蛋白、铁蛋白 磷脂酰胆碱、胆汁酸途径、烟酸和烟酰胺途径、磷脂酰肌醇和三羧酸循环 [57] 血浆 NAFLD组(n=427) 5-羟廿碳四烯酸,7,17-二氢吡啶二羧酸,肾上腺酸,花生四烯酸,二十碳五烯酸,16-羟基二十二碳六烯酸,9-羟基十八碳二烯酸 [58] 血清 NAFLD组(n=627) 2-羟基丁酸、3-羟基丁酸、柠檬酸、异亮氨酸、赖氨酸、油酸、3-OH-苯甲酸、5-OH-1H-吲哚-3-乙酸、吲哚-3-乳酸 [59] 尿液 NASH组(男68例,女65例) 高丙二酸乙酯、β-羟基丁酸、乙基丙二酸酯、硫酸盐水平、高甲氨基谷氨酸、对羟苯乳酸、琥珀酸盐、甲酰亚胺谷氨酸酯、香草扁桃酸酯、吡啶甲酸酯 酪氨酸分解代谢 [60] 尿液 NAFLD组(n=33),NASH组(n=45),健康对照组(n=30) 氨基酸代谢物、瓜氨酸、精氨酸、缬氨酸、吲哚乙酸以及葡萄糖和葡萄糖酸、次黄嘌呤、黄嘌呤和肉碱 脂质过氧化和氧化应激、氨基酸代谢和戊糖磷酸途径 [61] -
[1] ALBILLOS A, de GOTTARDI A, RESCIGNO M. The gut-liver axis in liver disease: Pathophysiological basis for therapy[J]. J Hepatol, 2020, 72( 3): 558- 577. DOI: 10.1016/j.jhep.2019.10.003. [2] ZENG FL, SHI MJ, XIAO HM, et al. WGCNA-based identification of hub genes and key pathways involved in nonalcoholic fatty liver disease[J]. Biomed Res Int, 2021, 2021: 5633211. DOI: 10.1155/2021/5633211. [3] ZENG TF, CHEN GL, QIAO XB, et al. NUSAP1 could be a potential target for preventing NAFLD progression to liver cancer[J]. Front Pharmacol, 2022, 13: 823140. DOI: 10.3389/fphar.2022.823140. [4] DAI WR, SUN Y, JIANG ZY, et al. Key genes associated with non-alcoholic fatty liver disease and acute myocardial infarction[J]. Med Sci Monit, 2020, 26: e922492. DOI: 10.12659/MSM.922492. [5] HANDELMAN SK, PUENTES YM, KUPPA A, et al. Population-based meta-analysis and gene-set enrichment identifies FXR/RXR pathway as common to fatty liver disease and serum lipids[J]. Hepatol Commun, 2022, 6( 11): 3120- 3131. DOI: 10.1002/hep4.2066. [6] CHEN JH, ZHOU H, JIN HW, et al. Role of inflammatory factors in mediating the effect of lipids on nonalcoholic fatty liver disease: A two-step, multivariable Mendelian randomization study[J]. Nutrients, 2022, 14( 20): 4434. DOI: 10.3390/nu14204434. [7] GHODSIAN N, ABNER E, EMDIN CA, et al. Electronic health record-based genome-wide meta-analysis provides insights on the genetic architecture of non-alcoholic fatty liver disease[J]. Cell Rep Med, 2021, 2( 11): 100437. DOI: 10.1016/j.xcrm.2021.100437. [8] SHARMA D, MANDAL P. NAFLD: Genetics and its clinical implications[J]. Clin Res Hepatol Gastroenterol, 2022, 46( 9): 102003. DOI: 10.1016/j.clinre.2022.102003. [9] NANO J, GHANBARI M, WANG WS, et al. Epigenome-wide association study identifies methylation sites associated with liver enzymes and hepatic steatosis[J]. Gastroenterology, 2017, 153( 4): 1096- 1106. e 2. DOI: 10.1053/j.gastro.2017.06.003. [10] ZHANG RN, PAN Q, ZHENG RD, et al. Genome-wide analysis of DNA methylation in human peripheral leukocytes identifies potential biomarkers of nonalcoholic fatty liver disease[J]. Int J Mol Med, 2018, 42( 1): 443- 452. DOI: 10.3892/ijmm.2018.3583. [11] WU JY, ZHANG RN, SHEN F, et al. Altered DNA methylation sites in peripheral blood leukocytes from patients with simple steatosis and nonalcoholic steatohepatitis(NASH)[J]. Med Sci Monit, 2018, 24: 6946- 6967. DOI: 10.12659/MSM.909747. [12] HYUN J, JUNG Y. DNA methylation in nonalcoholic fatty liver disease[J]. Int J Mol Sci, 2020, 21( 21): 8138. DOI: 10.3390/ijms21218138. [13] MA JT, NANO J, DING JZ, et al. A peripheral blood DNA methylation signature of hepatic fat reveals a potential causal pathway for nonalcoholic fatty liver disease[J]. Diabetes, 2019, 68( 5): 1073- 1083. DOI: 10.2337/DB18-1193. [14] ASSANTE G, CHANDRASEKARAN S, NG S, et al. Correction: Acetyl-CoA metabolism drives epigenome change and contributes to carcinogenesis risk in fatty liver disease[J]. Genome Med, 2023, 15( 1): 38. DOI: 10.1186/s13073-023-01190-7. [15] FU SF, YU MH, TAN YY, et al. Role of histone deacetylase on nonalcoholic fatty liver disease[J]. Expert Rev Gastroenterol Hepatol, 2021, 15( 4): 353- 361. DOI: 10.1080/17474124.2021.1854089. [16] CHUNG MY, KIM HJ, CHOI HK, et al. Black mulberry extract elicits hepatoprotective effects in nonalcoholic fatty liver disease models by inhibition of histone acetylation[J]. J Med Food, 2021, 24( 9): 978- 986. DOI: 10.1089/jmf.2021.K.0048. [17] BRICAMBERT J, ALVES-GUERRA MC, ESTEVES P, et al. The histone demethylase Phf2 acts as a molecular checkpoint to prevent NAFLD progression during obesity[J]. Nat Commun, 2018, 9: 2092. DOI: 10.1038/s41467-018-04361-y. [18] TIAN C, MIN XW, ZHAO YX, et al. MRG15 aggravates non-alcoholic steatohepatitis progression by regulating the mitochondrial proteolytic degradation of TUFM[J]. J Hepatol, 2022, 77( 6): 1491- 1503. DOI: 10.1016/j.jhep.2022.07.017. [19] CAO YN, XUE Y, XUE L, et al. Hepatic menin recruits SIRT1 to control liver steatosis through histone deacetylation[J]. J Hepatol, 2013, 59( 6): 1299- 1306. DOI: 10.1016/j.jhep.2013.07.011. [20] RIEGL SD, STARNES C, JIMA DD, et al. The imprinted gene Zac1 regulates steatosis in developmental cadmium-induced nonalcoholic fatty liver disease[J]. Toxicol Sci, 2023, 191( 1): 34- 46. DOI: 10.1093/toxsci/kfac106. [21] BAPTISSART M, BRADISH CM, JONES BS, et al. Zac1 and the Imprinted Gene Network program juvenile NAFLD in response to maternal metabolic syndrome[J]. Hepatology, 2022, 76( 4): 1090- 1104. DOI: 10.1002/hep.32363. [22] OKAMOTO K, KODA M, OKAMOTO T, et al. A series of microRNA in the chromosome 14q32.2 maternally imprinted region related to progression of non-alcoholic fatty liver disease in a mouse model[J]. PLoS One, 2016, 11( 5): e0154676. DOI: 10.1371/journal.pone.0154676. [23] ZHOU B, JIA LJ, ZHANG ZJ, et al. The nuclear orphan receptor NR2F6 promotes hepatic steatosis through upregulation of fatty acid transporter CD36[J]. Adv Sci(Weinh), 2020, 7( 21): 2002273. DOI: 10.1002/advs.202002273. [24] SUN CZ, LIU XY, YI ZJ, et al. Genome-wide analysis of long noncoding RNA expression profiles in patients with non-alcoholic fatty liver disease[J]. IUBMB Life, 2015, 67( 11): 847- 852. DOI: 10.1002/iub.1442. [25] LONG JK, DAI W, ZHENG YW, et al. MiR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease[J]. Mol Med, 2019, 25( 1): 26. DOI: 10.1186/s10020-019-0085-2. [26] GUO Y, XIONG YH, SHENG Q, et al. A micro-RNA expression signature for human NAFLD progression[J]. J Gastroenterol, 2016, 51( 10): 1022- 1030. DOI: 10.1007/s00535-016-1178-0. [27] HU MJ, LONG M, DAI RJ. Acetylation of H3K27 activated lncRNA NEAT1 and promoted hepatic lipid accumulation in non-alcoholic fatty liver disease via regulating miR-212-5p/GRIA3[J]. Mol Cell Biochem, 2022, 477( 1): 191- 203. DOI: 10.1007/s11010-021-04269-0. [28] OKAMOTO K, KODA M, OKAMOTO T, et al. Serum miR-379 expression is related to the development and progression of hypercholesterolemia in non-alcoholic fatty liver disease[J]. PLoS One, 2020, 15( 2): e0219412. DOI: 10.1371/journal.pone.0219412. [29] FANG ZQ, DOU GR, WANG L. MicroRNAs in the pathogenesis of nonalcoholic fatty liver disease[J]. Int J Biol Sci, 2021, 17( 7): 1851- 1863. DOI: 10.7150/ijbs.59588. [30] SHEN X, ZHANG YJ, JI XT, et al. Long noncoding RNA lncRHL regulates hepatic VLDL secretion by modulating hnRNPU/BMAL1/MTTP axis[J]. Diabetes, 2022, 71( 9): 1915- 1928. DOI: 10.2337/db21-1145. [31] JIN SS, LIN CJ, LIN XF, et al. Silencing lncRNA NEAT1 reduces nonalcoholic fatty liver fat deposition by regulating the miR-139-5p/c-Jun/SREBP-1c pathway[J]. Ann Hepatol, 2022, 27( 2): 100584. DOI: 10.1016/j.aohep.2021.100584. [32] ZUO ZH, ZENG CY, JIANG Y, et al. Regulatory role of long non-coding RNAs in the development and progression of nonalcoholic fatty liver disease[J]. J Clin Hepatol, 2021, 37( 7): 1704- 1707. DOI: 10.3969/j.issn.1001-5256.2021.07.048.左志华, 曾楚怡, 姜瑶, 等. 长链非编码RNA在非酒精性脂肪性肝病发生发展中的调控作用[J]. 临床肝胆病杂志, 2021, 37( 7): 1704- 1707. DOI: 10.3969/j.issn.1001-5256.2021.07.048. [33] YUAN XL, WANG J, TANG XY, et al. Berberine ameliorates nonalcoholic fatty liver disease by a global modulation of hepatic mRNA and lncRNA expression profiles[J]. J Transl Med, 2015, 13: 24. DOI: 10.1186/s12967-015-0383-6. [34] LI PF, SHAN KS, LIU Y, et al. CircScd1 promotes fatty liver disease via the Janus kinase 2/signal transducer and activator of transcription 5 pathway[J]. Dig Dis Sci, 2019, 64( 1): 113- 122. DOI: 10.1007/s10620-018-5290-2. [35] GUO XY, HE CX, WANG YQ, et al. Circular RNA profiling and bioinformatic modeling identify its regulatory role in hepatic steatosis[J]. Biomed Res Int, 2017, 2017: 5936171. DOI: 10.1155/2017/5936171. [36] CHEN X, TAN QQ, TAN XR, et al. Circ_0057558 promotes nonalcoholic fatty liver disease by regulating ROCK1/AMPK signaling through targeting miR-206[J]. Cell Death Dis, 2021, 12( 9): 809. DOI: 10.1038/s41419-021-04090-z. [37] LIU W, CAO HC, YAN J, et al.‘Micro-managers’ of hepatic lipid metabolism and NAFLD[J]. Wiley Interdiscip Rev RNA, 2015, 6( 5): 581- 593. DOI: 10.1002/wrna.1295. [38] HORIE T, NISHINO T, BABA O, et al. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice[J]. Nat Commun, 2013, 4: 2883. DOI: 10.1038/ncomms3883. [39] GOEDEKE L, SALERNO A, RAMÍREZ CM, et al. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice[J]. EMBO Mol Med, 2014, 6( 9): 1133- 1141. DOI: 10.15252/emmm.201404046. [40] CHEN Y, CHEN XY, GAO JG, et al. Long noncoding RNA FLRL2 alleviated nonalcoholic fatty liver disease through Arntl-Sirt1 pathway[J]. FASEB J, 2019, 33( 10): 11411- 11419. DOI: 10.1096/fj.201900643RRR. [41] ZAIOU M. Noncoding RNAs as additional mediators of epigenetic regulation in nonalcoholic fatty liver disease[J]. World J Gastroenterol, 2022, 28( 35): 5111- 5128. DOI: 10.3748/wjg.v28.i35.5111. [42] GONG ZH, TANG JL, XIANG TX, et al. Genome-wide identification of long noncoding RNAs in CCl4-induced liver fibrosis via RNA sequencing[J]. Mol Med Rep, 2018, 18( 1): 299- 307. DOI: 10.3892/mmr.2018.8986. [43] CHIEN Y, TSAI PH, LAI YH, et al. CircularRNA as novel biomarkers in liver diseases[J]. J Chin Med Assoc, 2020, 83( 1): 15- 17. DOI: 10.1097/JCMA.0000000000000230. [44] LI J, QI J, TANG YS, et al. A nanodrug system overexpressed circRNA_0001805 alleviates nonalcoholic fatty liver disease via miR-106a-5p/miR-320a and ABCA1/CPT1 axis[J]. J Nanobiotechnology, 2021, 19( 1): 363. DOI: 10.1186/s12951-021-01108-8. [45] JIN X, GAO JG, ZHENG RH, et al. Antagonizing circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy[J]. Cell Death Dis, 2020, 11( 2): 123. DOI: 10.1038/s41419-020-2293-7. [46] ZHANG LQ, ZHANG ZG, LI CB, et al. S100A11 promotes liver steatosis via FOXO1-mediated autophagy and lipogenesis[J]. Cell Mol Gastroenterol Hepatol, 2021, 11( 3): 697- 724. DOI: 10.1016/j.jcmgh.2020.10.006. [47] PENG YM, ZENG Q, WAN LM, et al. GP73 is a TBC-domain Rab GTPase-activating protein contributing to the pathogenesis of non-alcoholic fatty liver disease without obesity[J]. Nat Commun, 2021, 12( 1): 7004. DOI: 10.1038/s41467-021-27309-1. [48] LI JY, KOU CJ, SUN TT, et al. Identification and validation of hub immune-related genes in non-alcoholic fatty liver disease[J]. Int J Gen Med, 2023, 16: 2609- 2621. DOI: 10.2147/IJGM.S413545. [49] NIU LL, GEYER PE, WEWER ALBRECHTSEN NJ, et al. Plasma proteome profiling discovers novel proteins associated with non-alcoholic fatty liver disease[J]. Mol Syst Biol, 2019, 15( 3): e8793. DOI: 10.15252/msb.20188793. [50] da SILVA LIMA N, FONDEVILA MF, NÓVOA E, et al. Inhibition of ATG3 ameliorates liver steatosis by increasing mitochondrial function[J]. J Hepatol, 2022, 76( 1): 11- 24. DOI: 10.1016/j.jhep.2021.09.008. [51] YOKOYAMA C, WANG X, BRIGGS MR, et al. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene[J]. Cell, 1993, 75( 1): 187- 197. [52] WANG CE, XU WT, GONG J, et al. Research progress in treatment of nonalcoholic fatty liver disease[J]. Clin J Med Off, 2022, 50( 9): 897- 899, 903. DOI: 10.16680/j.1671-3826.2022.09.06.王彩娥, 许文涛, 宫建, 等. 非酒精性脂肪性肝病治疗研究进展[J]. 临床军医杂志, 2022, 50( 9): 897- 899, 903. DOI: 10.16680/j.1671-3826.2022.09.06. [53] JUNG Y, LEE MK, PURI P, et al. Circulating lipidomic alterations in obese and non-obese subjects with non-alcoholic fatty liver disease[J]. Aliment Pharmacol Ther, 2020, 52( 10): 1603- 1614. DOI: 10.1111/apt.16066. [54] LU QR, TIAN XY, WU H, et al. Metabolic changes of hepatocytes in NAFLD[J]. Front Physiol, 2021, 12: 710420. DOI: 10.3389/fphys.2021.710420. [55] CANFORA EE, MEEX RCR, VENEMA K, et al. Gut microbial metabolites in obesity, NAFLD and T2DM[J]. Nat Rev Endocrinol, 2019, 15( 5): 261- 273. DOI: 10.1038/s41574-019-0156-z. [56] TANG Y, CHEN X, CHEN Q, et al. Association of serum methionine metabolites with non-alcoholic fatty liver disease: A cross-sectional study[J]. Nutr Metab, 2022, 19( 1): 21. DOI: 10.1186/s12986-022-00647-7. [57] OGAWA Y, KOBAYASHI T, HONDA Y, et al. Metabolomic/lipidomic-based analysis of plasma to diagnose hepatocellular ballooning in patients with non-alcoholic fatty liver disease: A multicenter study[J]. Hepatol Res, 2020, 50( 8): 955- 965. DOI: 10.1111/hepr.13528. [58] CAUSSY C, CHUANG JC, BILLIN A, et al. Plasma eicosanoids as noninvasive biomarkers of liver fibrosis in patients with nonalcoholic steatohepatitis[J]. Therap Adv Gastroenterol, 2020, 13: 1756284820923904. DOI: 10.1177/1756284820923904. [59] MCGLINCHEY AJ, GOVAERE O, GENG DW, et al. Metabolic signatures across the full spectrum of non-alcoholic fatty liver disease[J]. JHEP Rep, 2022, 4( 5): 100477. DOI: 10.1016/j.jhepr.2022.100477. [60] HAAM JH, LEE YK, SUH E, et al. Characteristics of urine organic acid metabolites in nonalcoholic fatty liver disease assessed using magnetic resonance imaging with elastography in Korean adults[J]. Diagnostics(Basel), 2022, 12( 5): 1199. DOI: 10.3390/diagnostics12051199. [61] DONG S, ZHAN ZY, CAO HY, et al. Urinary metabolomics analysis identifies key biomarkers of different stages of nonalcoholic fatty liver disease[J]. World J Gastroenterol, 2017, 23( 15): 2771- 2784. DOI: 10.3748/wjg.v23.i15.2771. [62] KIM HY. Recent advances in nonalcoholic fatty liver disease metabolomics[J]. Clin Mol Hepatol, 2021, 27( 4): 553- 559. DOI: 10.3350/cmh.2021.0127. [63] HOU TY, TIAN Y, CAO ZY, et al. Cytoplasmic SIRT6-mediated ACSL5 deacetylation impedes nonalcoholic fatty liver disease by facilitating hepatic fatty acid oxidation[J]. Mol Cell, 2022, 82( 21): 4099- 4115. e 9. DOI: 10.1016/j.molcel.2022.09.018. [64] DONG QM, KUEFNER MS, DENG X, et al. Sex-specific differences in hepatic steatosis in obese spontaneously hypertensive(SHROB) rats[J]. Biol Sex Differ, 2018, 9( 1): 40. DOI: 10.1186/s13293-018-0202-x. [65] LIU J, SHI Y, PENG DY, et al. Salvia miltiorrhiza bge.(Danshen) in the treating non-alcoholic fatty liver disease based on the regulator of metabolic targets[J]. Front Cardiovasc Med, 2022, 9: 842980. DOI: 10.3389/fcvm.2022.842980. [66] di CIAULA A, PASSARELLA S, SHANMUGAM H, et al. Nonalcoholic fatty liver disease(NAFLD). mitochondria as players and targets of therapies?[J]. Int J Mol Sci, 2021, 22( 10): 5375. DOI: 10.3390/ijms22105375. [67] ESLER WP, BENCE KK. Metabolic targets in nonalcoholic fatty liver disease[J]. Cell Mol Gastroenterol Hepatol, 2019, 8( 2): 247- 267. DOI: 10.1016/j.jcmgh.2019.04.007. [68] CHEN JZ, VITETTA L. Gut microbiota metabolites in NAFLD pathogenesis and therapeutic implications[J]. Int J Mol Sci, 2020, 21( 15): 5214. DOI: 10.3390/ijms21155214. -



 PDF下载 ( 944 KB)
PDF下载 ( 944 KB)

 下载:
下载:


