| [1] |
MODY K, ABOU-ALFA GK. Systemic therapy for advanced hepatocellular carcinoma in an evolving landscape[J]. Curr Treat Options Oncol, 2019, 20( 2): 3. DOI: 10.1007/s11864-019-0601-1. |
| [2] |
BRAY F, FERLAY J, SOERJOMATARAM I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68( 6): 394- 424. DOI: 10.3322/caac.21492. |
| [3] |
SCHLABE S, ROCKSTROH JK. Advances in the treatment of HIV/HCV coinfection in adults[J]. Expert Opin Pharmacother, 2018, 19( 1): 49- 64. DOI: 10.1080/14656566.2017.1419185. |
| [4] |
CHEN Z, XIE H, HU M, et al. Recent progress in treatment of hepatocellular carcinoma[J]. Am J Cancer Res, 2020, 10( 9): 2993- 3036.
|
| [5] |
NORDENSTEDT H, WHITE DL, EL-SERAG HB. The changing pattern of epidemiology in hepatocellular carcinoma[J]. Dig Liver Dis, 2010, 42( Suppl 3): S206- S214. DOI: 10.1016/S1590-8658(10)60507-5. |
| [6] |
SCHWEITZER A, HORN J, MIKOLAJCZYK RT, et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013[J]. Lancet, 2015, 386( 10003): 1546- 1555. DOI: 10.1016/S0140-6736(15)61412-X. |
| [7] |
WILD CP, MILLER JD, GROOPMAN JD, et al. Mycotoxin control in low-and middle-income countries[M]. Lyon(FR): International Agency for Research on Cancer © International Agency for Research on Cancer, 2015.
|
| [8] |
BARTENSCHLAGER R, SCHALLER H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome[J]. EMBO J, 1992, 11( 9): 3413- 3420. DOI: 10.1002/j.1460-2075.1992.tb05420.x. |
| [9] |
HIRSCH RC, LOEB DD, POLLACK JR, et al. cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA[J]. J Virol, 1991, 65( 6): 3309- 3316. DOI: 10.1128/JVI.65.6.3309-3316.1991. |
| [10] |
WEI L, PLOSS A. Hepatitis B virus cccDNA is formed through distinct repair processes of each strand[J]. Nat Commun, 2021, 12( 1): 1591. DOI: 10.1038/s41467-021-21850-9. |
| [11] |
SELZER L, ZLOTNICK A. Assembly and release of hepatitis B virus[J]. Cold Spring Harb Perspect Med, 2015, 5( 12): a021394. DOI: 10.1101/cshperspect.a021394. |
| [12] |
ZLOTNICK A, VENKATAKRISHNAN B, TAN Z, et al. Core protein: A pleiotropic keystone in the HBV lifecycle[J]. Antiviral Res, 2015, 121: 82- 93. DOI: 10.1016/j.antiviral.2015.06.020. |
| [13] |
INOUE T, TANAKA Y. The Role of hepatitis B core-related antigen[J]. Genes(Basel), 2019, 10( 5): 357. DOI: 10.3390/genes10050357. |
| [14] |
|
| [15] |
BENHENDA S, COUGOT D, BUENDIA MA, et al. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis[J]. Adv Cancer Res, 2009, 103: 75- 109. DOI: 10.1016/S0065-230X(09)03004-8. |
| [16] |
SLAGLE BL, BOUCHARD MJ. Role of HBx in hepatitis B virus persistence and its therapeutic implications[J]. Curr Opin Virol, 2018, 30: 32- 38. DOI: 10.1016/j.coviro.2018.01.007. |
| [17] |
BELLONI L, POLLICINO T, de NICOLA F, et al. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function[J]. Proc Natl Acad Sci U S A, 2009, 106( 47): 19975- 19979. DOI: 10.1073/pnas.0908365106. |
| [18] |
HU J, FLORES D, TOFT D, et al. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function[J]. J Virol, 2004, 78( 23): 13122- 13131. DOI: 10.1128/JVI.78.23.13122-13131.2004. |
| [19] |
KIM S, WANG H, RYU WS. Incorporation of eukaryotic translation initiation factor eIF4E into viral nucleocapsids via interaction with hepatitis B virus polymerase[J]. J Virol, 2010, 84( 1): 52- 58. DOI: 10.1128/JVI.01232-09. |
| [20] |
JONES SA, HU J. Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging targets for antiviral intervention[J]. Emerg Microbes Infect, 2013, 2( 9): e56. DOI: 10.1038/emi.2013.56. |
| [21] |
YU S, CHEN J, WU M, et al. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKepsilon and DDX3[J]. J Gen Virol, 2010, 91( Pt 8): 2080- 2090. DOI: 10.1099/vir.0.020552-0. |
| [22] |
WANG H, RYU WS. Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: implications for immune evasion[J]. PLoS Pathog, 2010, 6( 7): e1000986. DOI: 10.1371/journal.ppat.1000986. |
| [23] |
LIU Y, LI J, CHEN J, et al. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways[J]. J Virol, 2015, 89( 4): 2287- 2300. DOI: 10.1128/JVI.02760-14. |
| [24] |
SUN Y, YU M, QU M, et al. Hepatitis B virus-triggered PTEN/β-catenin/c-Myc signaling enhances PD-L1 expression to promote immune evasion[J]. Am J Physiol Gastrointest Liver Physiol, 2020, 318( 1): G162- G173. DOI: 10.1152/ajpgi.00197.2019. |
| [25] |
LIU M, WU H, LIU T, et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma[J]. Cell Res, 2009, 19( 7): 828- 837. DOI: 10.1038/cr.2009.72. |
| [26] |
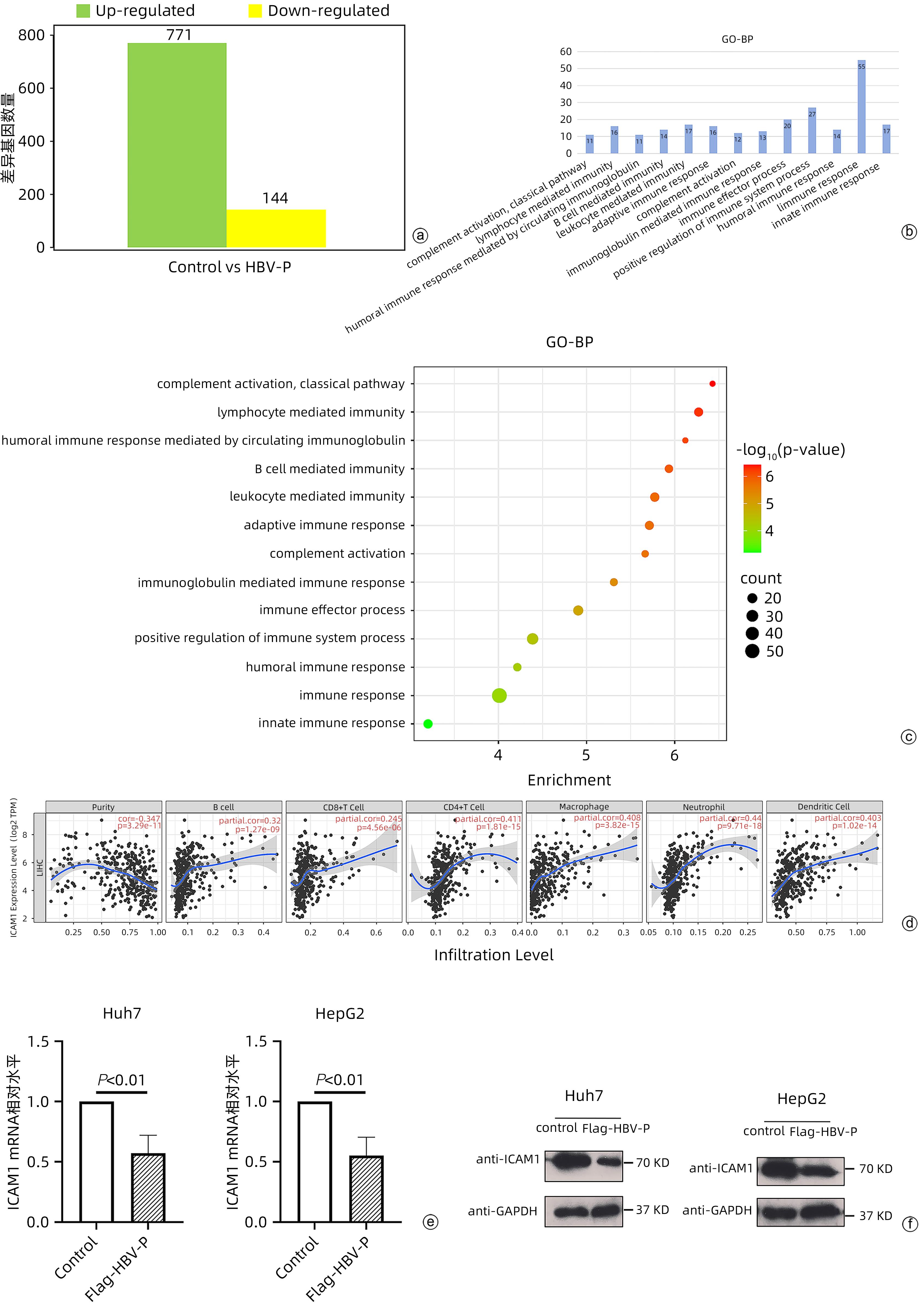
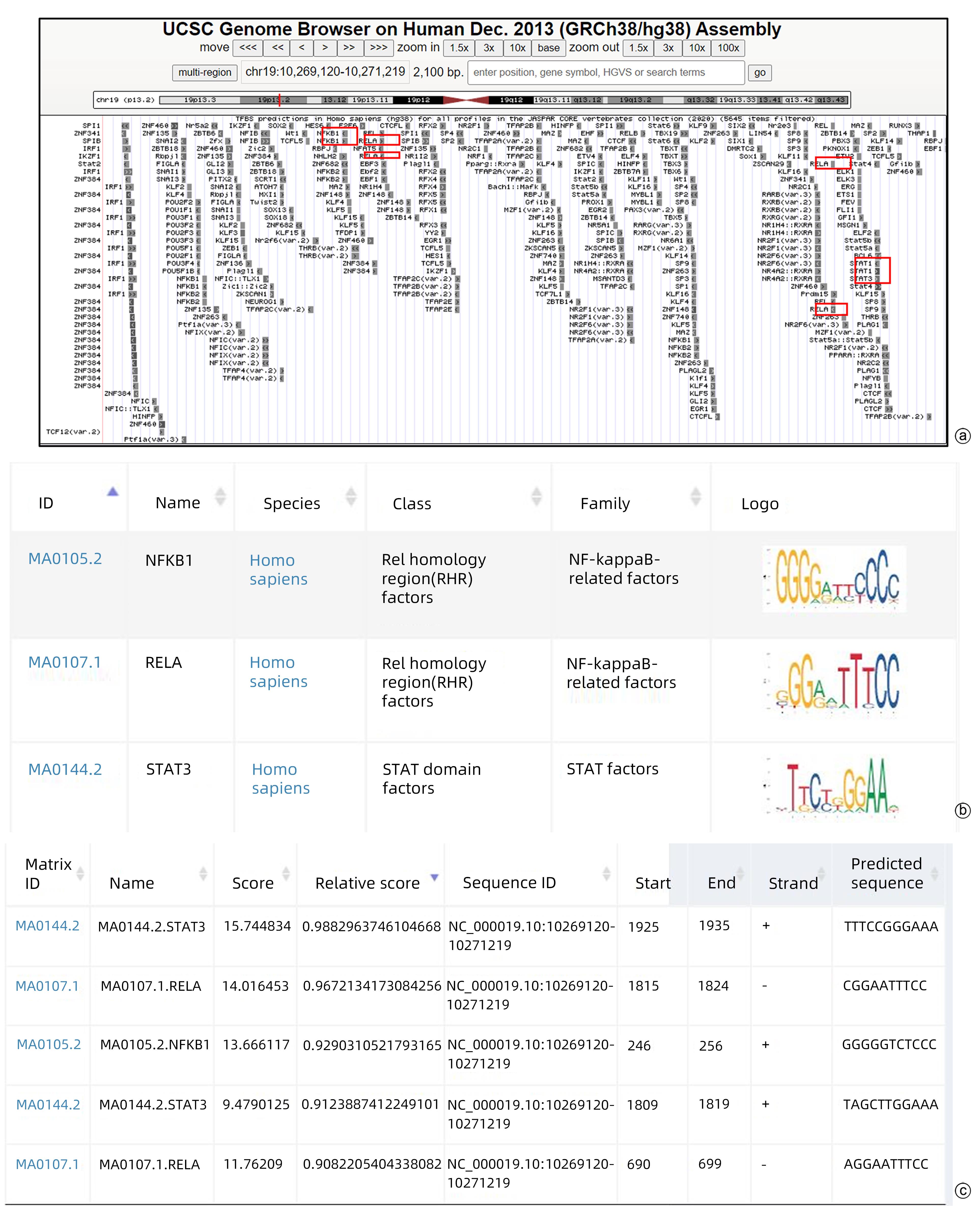
CHEN Y, XU Y, ZHAO M, et al. High-throughput T cell receptor sequencing reveals distinct repertoires between tumor and adjacent non-tumor tissues in HBV-associated HCC[J]. Oncoimmunology, 2016, 5( 10): e1219010. DOI: 10.1080/2162402X.2016.1219010. |
| [27] |
STAUNTON DE, MARLIN SD, STRATOWA C, et al. Primary structure of ICAM-1 demonstrates interaction between members of the immunoglobulin and integrin supergene families[J]. Cell, 1988, 52( 6): 925- 933. DOI: 10.1016/0092-8674(88)90434-5. |
| [28] |
HUBBARD AK, ROTHLEIN R. Intercellular adhesion molecule-1(ICAM-1) expression and cell signaling cascades[J]. Free Radic Biol Med, 2000, 28( 9): 1379- 1386. DOI: 10.1016/s0891-5849(00)00223-9. |
| [29] |
WEE H, OH HM, JO JH, et al. ICAM-1/LFA-1 interaction contributes to the induction of endothelial cell-cell separation: implication for enhanced leukocyte diapedesis[J]. Exp Mol Med, 2009, 41( 5): 341- 348. DOI: 10.3858/emm.2009.41.5.038. |
| [30] |
GORINA R, LYCK R, VESTWEBER D, et al. β2 integrin-mediated crawling on endothelial ICAM-1 and ICAM-2 is a prerequisite for transcellular neutrophil diapedesis across the inflamed blood-brain barrier[J]. J Immunol, 2014, 192( 1): 324- 337. DOI: 10.4049/jimmunol.1300858. |
| [31] |
BACHMANN MF, MCKALL-FAIENZA K, SCHMITS R, et al. Distinct roles for LFA-1 and CD28 during activation of naive T cells: adhesion versus costimulation[J]. Immunity, 1997, 7( 4): 549- 557. DOI: 10.1016/s1074-7613(00)80376-3. |
| [32] |
JENKINSON SR, WILLIAMS NA, MORGAN DJ. The role of intercellular adhesion molecule-1/LFA-1 interactions in the generation of tumor-specific CD8 + T cell responses[J]. J Immunol, 2005, 174( 6): 3401- 3407. DOI: 10.4049/jimmunol.174.6.3401. |
| [33] |
POGGI A, PREVOSTO C, ZANCOLLI M, et al. NKG2D and natural cytotoxicity receptors are involved in natural killer cell interaction with self-antigen presenting cells and stromal cells[J]. Ann N Y Acad Sci, 2007, 1109: 47- 57. DOI: 10.1196/annals.1398.007. |








 DownLoad:
DownLoad: