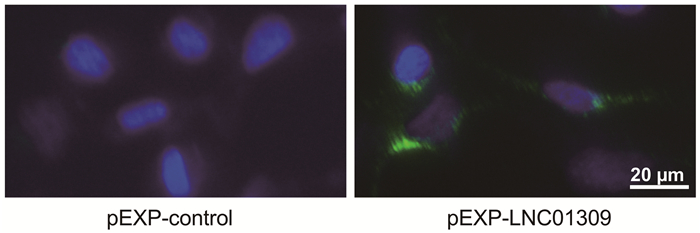
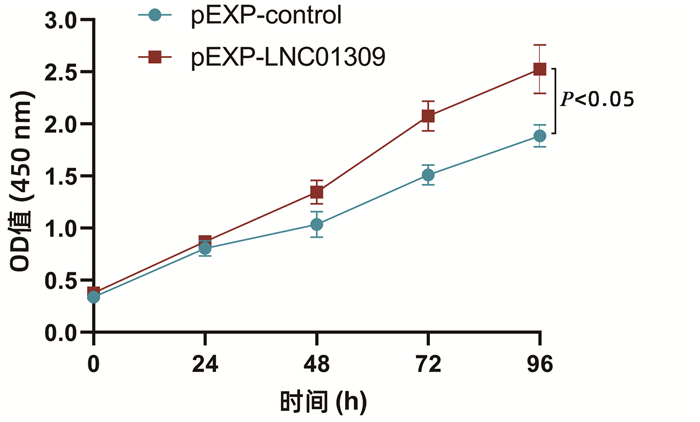
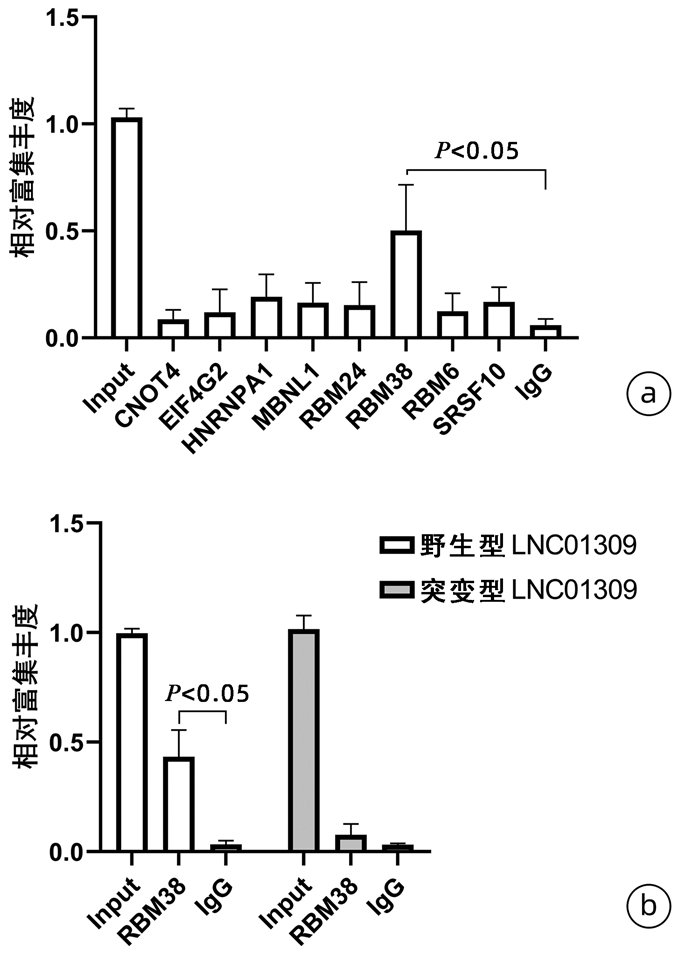
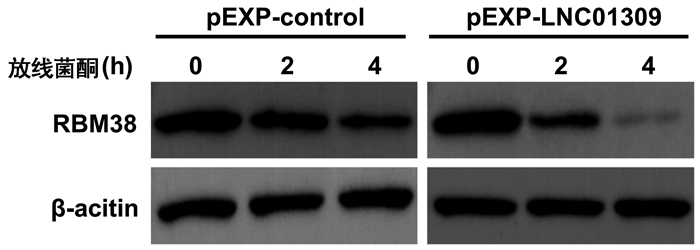
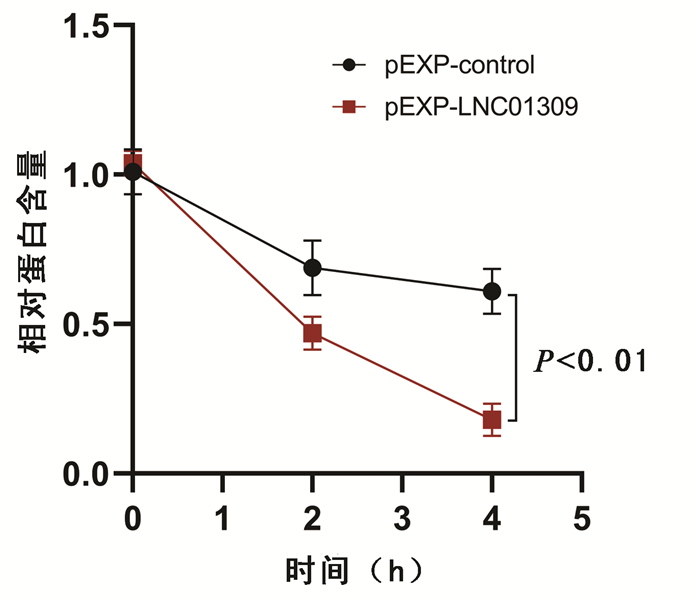
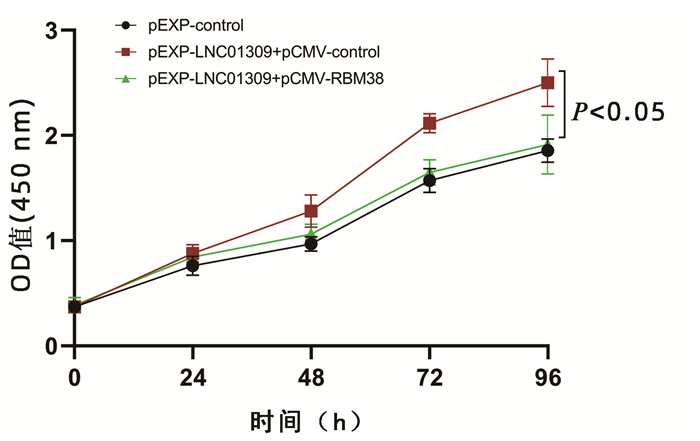
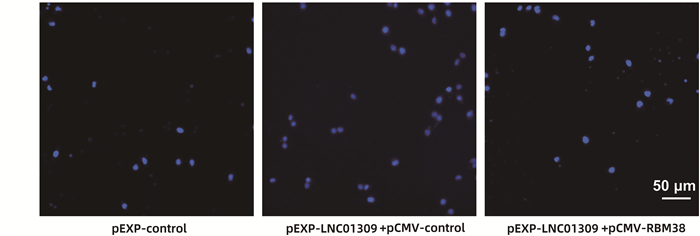
| [1] |
BHAN A, SOLEIMANI M, MANDAL SS. Long noncoding RNA and cancer: A new paradigm[J]. Cancer Res, 2017, 77(15): 3965-3981. DOI: 10.1158/0008-5472.CAN-16-2634. |
| [2] |
PENG WX, KOIRALA P, MO YY. LncRNA-mediated regulation of cell signaling in cancer[J]. Oncogene, 2017, 36(41): 5661-5667. DOI: 10.1038/onc.2017.184. |
| [3] |
|
| [4] |
KUMAR S, GONZALEZ EA, RAMESHWAR P, et al. Non-coding RNAs as mediators of epigenetic changes in malignancies[J]. Cancers (Basel), 2020, 12(12): 3657. DOI: 10.3390/cancers12123657. |
| [5] |
MA L, BAJIC VB, ZHANG Z. On the classification of long non-coding RNAs[J]. RNA Biol, 2013, 10(6): 925-933. DOI: 10.4161/rna.24604. |
| [6] |
MIYAMOTO M, MOTOOKA D, GOTOH K, et al. Performance comparison of second- and third-generation sequencers using a bacterial genome with two chromosomes[J]. BMC Genomics, 2014, 15: 699. DOI: 10.1186/1471-2164-15-699. |
| [7] |
RHOADS A, AU KF. PacBio sequencing and its applications[J]. Genomics Proteomics Bioinformatics, 2015, 13(5): 278-289. DOI: 10.1016/j.gpb.2015.08.002. |
| [8] |
XU J, ZHENG Y, PU S, et al. Third-generation sequencing found LncRNA associated with heat shock protein response to heat stress in Populus qiongdaoensis seedlings[J]. BMC Genomics, 2020, 21(1): 572. DOI: 10.1186/s12864-020-06979-z. |
| [9] |
SHEN Y, LIANG W, LIN Y, et al. Single molecule real-time sequencing and RNA-seq unravel the role of long non-coding and circular RNA in the regulatory network during Nile tilapia (Oreochromis niloticus) infection with Streptococcus agalactiae[J]. Fish Shellfish Immunol, 2020, 104: 640-653. DOI: 10.1016/j.fsi.2020.06.015. |
| [10] |
HUANG Z, ZHOU JK, PENG Y, et al. The role of long noncoding RNAs in hepatocellular carcinoma[J]. Mol Cancer, 2020, 19(1): 77. DOI: 10.1186/s12943-020-01188-4. |
| [11] |
CHEN Y, LI Z, CHEN X, et al. Long non-coding RNAs: From disease code to drug role[J]. Acta Pharm Sin B, 2021, 11(2): 340-354. DOI: 10.1016/j.apsb.2020.10.001. |
| [12] |
LI H, JI FX, XU XN, et al. Effects of long-chain non-coding RNA LINC00467 targeting miR-1247-5p on proliferation, migration and invasion of hepatocellular carcinoma cells[J]. Chin J Gerontol, 2021, 41(15): 3328-3333. DOI: 10.3969/j.issn.1005-9202.2021.15.048 |
| [13] |
YUAN JH, YANG F, WANG F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma[J]. Cancer Cell, 2014, 25(5): 666-681. DOI: 10.1016/j.ccr.2014.03.010. |
| [14] |
PANZITT K, TSCHERNATSCH MM, GUELLY C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA[J]. Gastroenterology, 2007, 132(1): 330-342. DOI: 10.1053/j.gastro.2006.08.026. |
| [15] |
DU Y, KONG G, YOU X, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18[J]. J Biol Chem, 2012, 287(31): 26302-26311. DOI: 10.1074/jbc.M112.342113. |
| [16] |
GENG YJ, XIE SL, LI Q, et al. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression[J]. J Int Med Res, 2011, 39(6): 2119-2128. DOI: 10.1177/147323001103900608. |
| [17] |
LI G, ZHANG H, WAN X, et al. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma[J]. Biomed Res Int, 2014, 2014: 780521. DOI: 10.1155/2014/780521. |
| [18] |
MAASS PG, LUFT FC, BÄHRING S. Long non-coding RNA in health and disease[J]. J Mol Med (Berl), 2014, 92(4): 337-346. DOI: 10.1007/s00109-014-1131-8. |
| [19] |
|
| [20] |
|
| [21] |
HUANG JZ, CHEN M, CHEN, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth[J]. Mol Cell, 2017, 68(1): 171-184. DOI: 10.1016/j.molcel.2017.09.015. |
| [22] |
LI XX, SHI L, ZHOU XJ, et al. The role of c-Myc-RBM38 loop in the growth suppression in breast cancer[J]. J Exp Clin Cancer Res, 2017, 36(1): 49. DOI: 10.1186/s13046-017-0521-5. |
| [23] |
JIANG Y, XU E, ZHANG J, et al. The Rbm38-p63 feedback loop is critical for tumor suppression and longevity[J]. Oncogene, 2018, 37(21): 2863-2872. DOI: 10.1038/s41388-018-0176-5. |
| [24] |
CHO SJ, ZHANG J, CHEN X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability[J]. Nucleic Acids Res, 2010, 38(7): 2256-2267. DOI: 10.1093/nar/gkp1229. |
| [25] |
XU E, ZHANG J, CHEN X. MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability[J]. Oncogene, 2013, 32(17): 2169-2178. DOI: 10.1038/onc.2012.238. |
| [26] |
YAN W, ZHANG J, ZHANG Y, et al. p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability[J]. Mol Cell Biol, 2012, 32(13): 2336-2348. DOI: 10.1128/MCB.00215-12. |
| [27] |
SUN Z, YANG GS, SIMA H, et al. Expression of RBM5 and its effect on prognosis of hepatocellular carcinoma patients after hepatectomy[J/CD]. Chin J Hepat Surg (Electronic Edition), 2021, 10(1): 93-97. DOI: 10.3877/cma.j.issn.2095-3232.2021.01.020. |
| [28] |
YE J, LIANG R, BAI T, et al. RBM38 plays a tumor-suppressor role via stabilizing the p53-mdm2 loop function in hepatocellular carcinoma[J]. J Exp Clin Cancer Res, 2018, 37(1): 212. DOI: 10.1186/s13046-018-0852-x. |



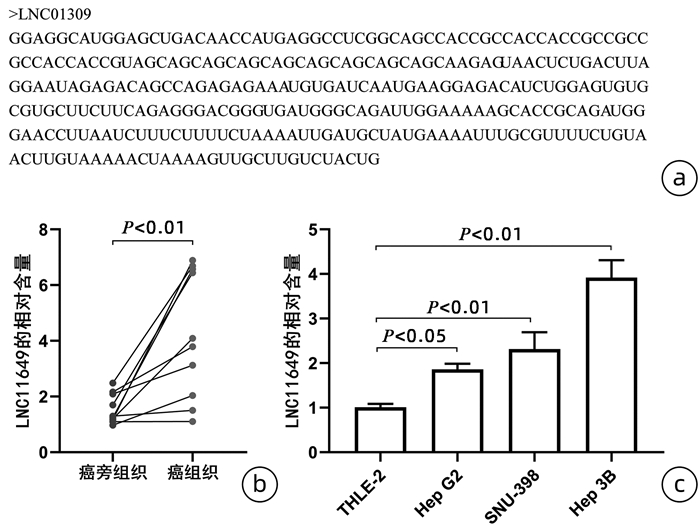




 DownLoad:
DownLoad: