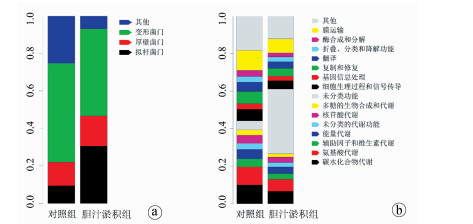
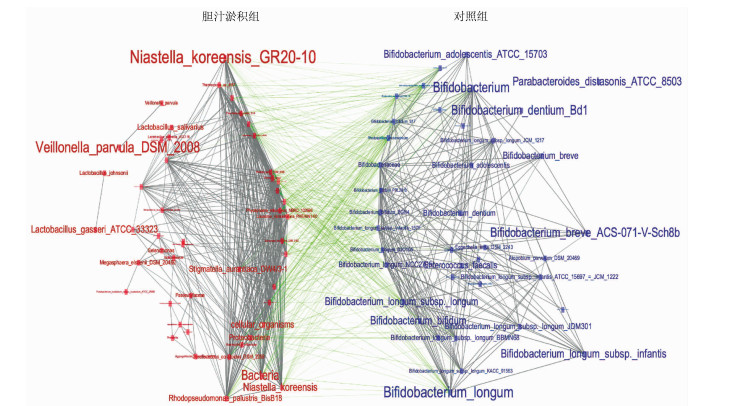
| [1] |
LIU Y, HUANG ZH. Clinical diagnosis and treatment of cholestatic hepatopathy in infants[J]. J Clin Hepatol, 2015, 31(8): 1218-1220. (in Chinese) DOI: 10.3969/j.issn.1001-5256.2015.08.009 |
| [2] |
FISCHLER B, LAMIREAU T. Cholestasis in the newborn and infant[J]. Clin Res Hepatol Gastroenterol, 2014, 38(3): 263-267. DOI: 10.1016/j.clinre.2014.03.010 |
| [3] |
CESARO C, TISO A, DEL PRETE A, et al. Gut microbiota and probiotics in chronic liver diseases[J]. Dig Liver Dis, 2011, 43(6): 431-438. DOI: 10.1016/j.dld.2010.10.015 |
| [4] |
TRAUNER M, FICKERT P, TILG H. Bile acids as modulators of gut microbiota linking dietary habits and inflammatory bowel disease: A potentially dangerous liaison[J]. Gastroenterology, 2013, 144(4): 844-846. DOI: 10.1053/j.gastro.2013.02.029 |
| [5] |
TRAUNER M, HALILBASIC E. Nuclear receptors as new perspective for the management of liver diseases[J]. Gastroenterology, 2011, 140(4): 1120-1125. e1-e12. DOI: 10.1053/j.gastro.2011.02.044 |
| [6] |
QIN N, YANG F, LI A, et al. Alterations of the human gut microbiome in liver cirrhosis[J]. Nature, 2014, 513(7516): 59-64. DOI: 10.1038/nature13568 |
| [7] |
ZHOU S, XU R, HE F, et al. Diversity of gut microbiota metabolic pathways in 10 pairs of Chinese infant twins[J]. PLoS One, 2016, 11(9): e0161627. DOI: 10.1371/journal.pone.0161627 |
| [8] |
HUSON DH, AUCH AF, QI J, et al. MEGAN analysis of metagenomic data[J]. Genome Res, 2007, 17(3): 377-386. DOI: 10.1101/gr.5969107 |
| [9] |
|
| [10] |
MANYAM G, BIRERDINC A, BARANOVA A. KPP: KEGG pathway painter[J]. BMC Syst Biol, 2015, 9(Suppl 2): s3. DOI: 10.1186/1752-0509-9-S2-S3 |
| [11] |
|
| [12] |
LOVE MI, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. DOI: 10.1186/s13059-014-0550-8 |
| [13] |
SHANNON P, MARKIEL A, OZIER O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks[J]. Genome Res, 2003, 13(11): 2498-2504. DOI: 10.1101/gr.1239303 |
| [14] |
HUANG Q, ZHANG HB, LI JT, et al. Research advances in the mechanism of action of intestinal microecology in intrahepatic cholestasis[J]. J Clin Hepatol, 2019, 35(10): 2355-2359. (in Chinese) DOI: 10.3969/j.issn.1001-5256.2019.10.050 |
| [15] |
RIDLON JM, ALVES JM, HYLEMON PB, et al. Cirrhosis, bile acids and gut microbiota: Unraveling a complex relationship[J]. Gut Microbes, 2013, 4(5): 382-387. DOI: 10.4161/gmic.25723 |
| [16] |
COSTELLO EK, STAGAMAN K, DETHLEFSEN L, et al. The application of ecological theory toward an understanding of the human microbiome[J]. Science, 2012, 336(6086): 1255-1262. DOI: 10.1126/science.1224203 |
| [17] |
TROSVIK P, STENSETH NC, RUDI K. Convergent temporal dynamics of the human infant gut microbiota[J]. ISME J, 2010, 4(2): 151-158. DOI: 10.1038/ismej.2009.96 |
| [18] |
|
| [19] |
OLSZAK T, AN D, ZEISSIG S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function[J]. Science, 2012, 336(6080): 489-493. DOI: 10.1126/science.1219328 |
| [20] |
CHEN Y, YANG F, LU H, et al. Characterization of fecal microbial communities in patients with liver cirrhosis[J]. Hepatology, 2011, 54(2): 562-572. DOI: 10.1002/hep.24423 |
| [21] |
|
| [22] |
BÄCKHED F, ROSWALL J, PENG Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life[J]. Cell Host Microbe, 2015, 17(6): 852. DOI: 10.1016/j.chom.2015.05.012 |
| [23] |
BAJAJ JS, RIDLON JM, HYLEMON PB, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy[J]. Am J Physiol Gastrointest Liver Physiol, 2012, 302(1): g168-g175. DOI: 10.1152/ajpgi.00190.2011 |
| [24] |
FUKUI H. Gut-liver axis in liver cirrhosis: How to manage leaky gut and endotoxemia[J]. World J Hepatol, 2015, 7(3): 425-442. DOI: 10.4254/wjh.v7.i3.425 |
| [25] |
BAUER TM, SCHWACHA H, STEINBRVCKNER B, et al. Small intestinal bacterial overgrowth in human cirrhosis is associated with systemic endotoxemia[J]. Am J Gastroenterol, 2002, 97(9): 2364-2370. DOI: 10.1111/j.1572-0241.2002.05791.x |
| [26] |
|
| [27] |
QIN J, LI Y, CAI Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes[J]. Nature, 2012, 490(7418): 55-60. DOI: 10.1038/nature11450 |
| [28] |
GREENBLUM S, TURNBAUGH PJ, BORENSTEIN E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease[J]. Proc Natl Acad Sci U S A, 2012, 109(2): 594-599. DOI: 10.1073/pnas.1116053109 |
| [29] |
LEPAGE P, HÄSLER R, SPEHLMANN ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis[J]. Gastroenterology, 2011, 141(1): 227-236. DOI: 10.1053/j.gastro.2011.04.011 |



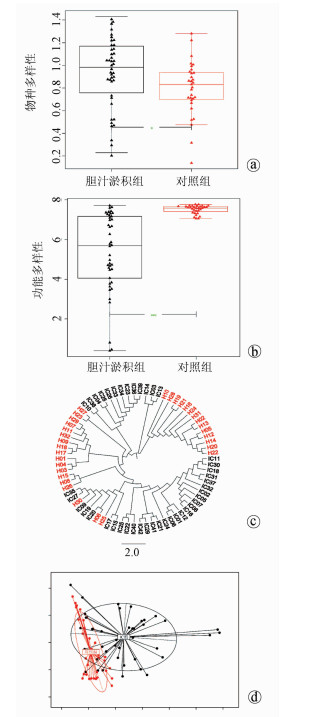




 DownLoad:
DownLoad: