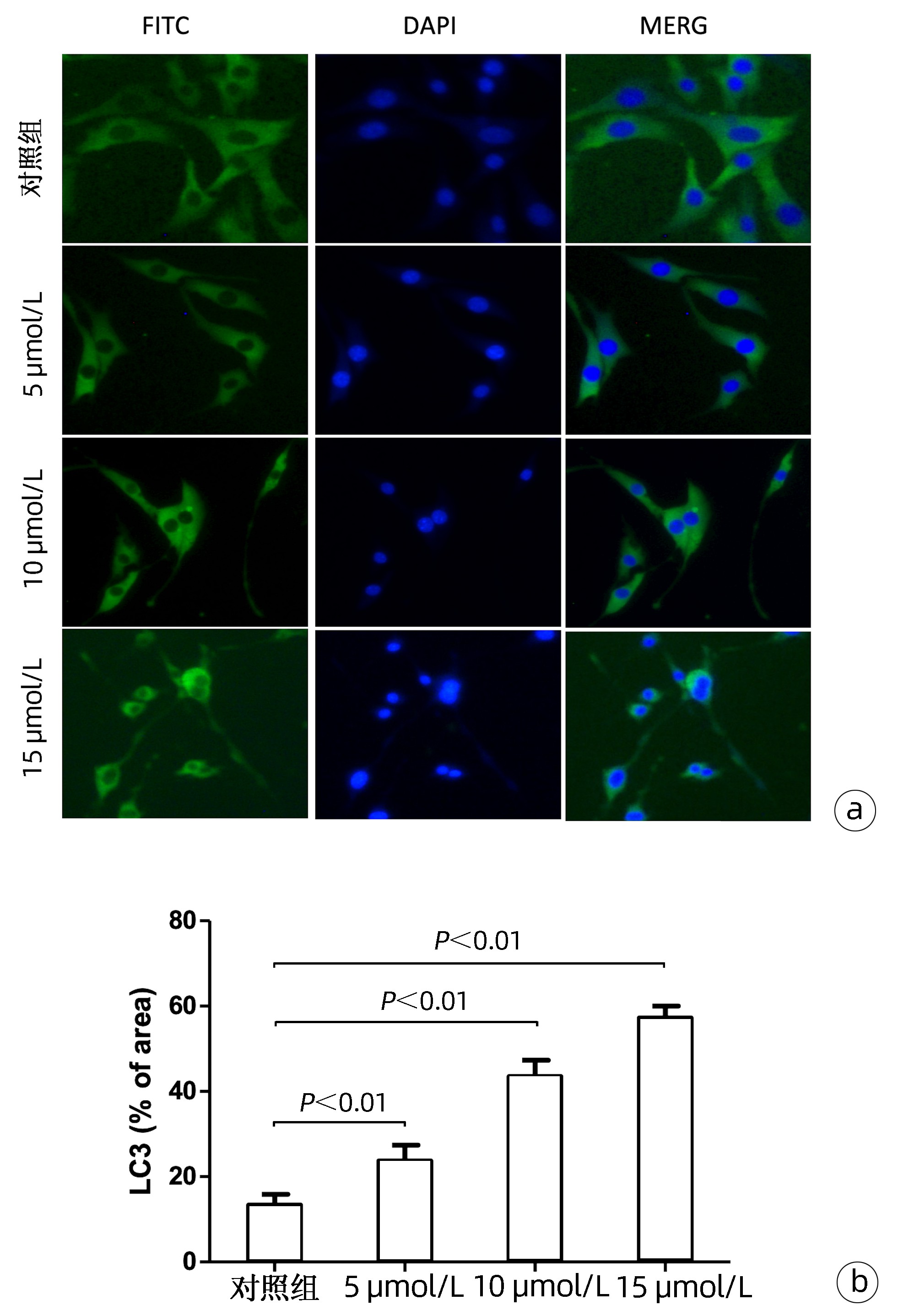
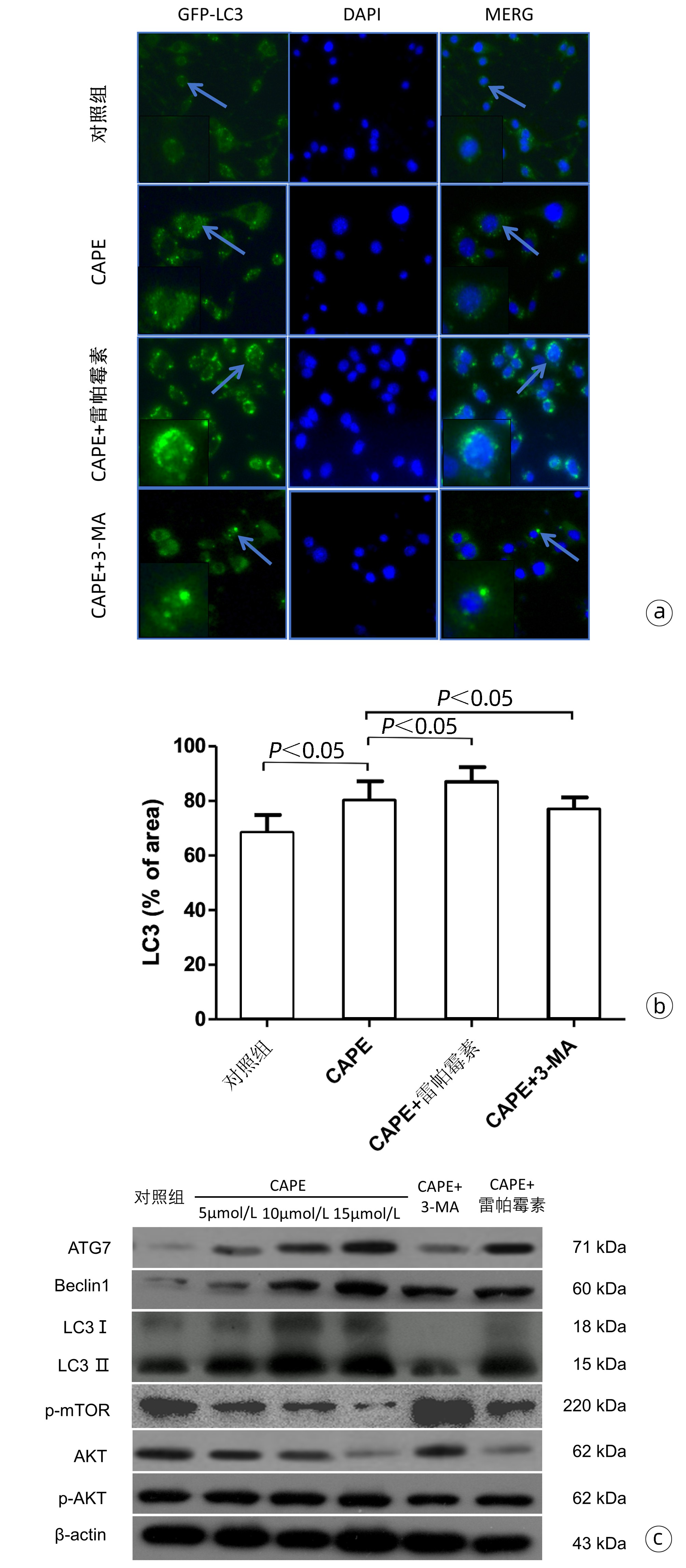
| [1] |
MURTAZA G, KARIM S, AKRAM MR, et al. Caffeic acid phenethyl ester and therapeutic potentials[J]. Biomed Res Int, 2014, 2014: 145342. DOI: 10.1155/2014/145342. |
| [2] |
OLGIERD B, KAMILA Z, ANNA B, et al. The pluripotent activities of caffeic acid phenethyl ester[J]. Molecules, 2021, 26(5): 1335. DOI: 10.3390/molecules26051335. |
| [3] |
LI M, WANG XF, SHI JJ, et al. Caffeic acid phenethyl ester inhibits liver fibrosis in rats[J]. World J Gastroenterol, 2015, 21(13): 3893-3903. DOI: 10.3748/wjg.v21.i13.3893. |
| [4] |
YANG N, SHI JJ, WU FP, et al. Caffeic acid phenethyl ester up-regulates antioxidant levels in hepatic stellate cell line T6 via an Nrf2-mediated mitogen activated protein kinases pathway[J]. World J Gastroenterol, 2017, 23(7): 1203-1214. DOI: 10.3748/wjg.v23.i7.1203. |
| [5] |
HAZARI Y, BRAVO-SAN PEDRO JM, HETZ C, et al. Autophagy in hepatic adaptation to stress[J]. J Hepatol, 2020, 72(1): 183-196. DOI: 10.1016/j.jhep.2019.08.026. |
| [6] |
KOUROUMALIS E, VOUMVOURAKI A, AUGOUSTAKI A, et al. Autophagy in liver diseases[J]. World J Hepatol, 2021, 13(1): 6-65. DOI: 10.4254/wjh.v13.i1.6. |
| [7] |
ALLAIRE M, RAUTOU PE, CODOGNO P, et al. Autophagy in liver diseases: Time for translation?[J]. J Hepatol, 2019, 70(5): 985-998. DOI: 10.1016/j.jhep.2019.01.026. |
| [8] |
ROEHLEN N, CROUCHET E, BAUMERT TF. Liver fibrosis: mechanistic concepts and therapeutic perspectives[J]. Cells, 2020, 9(4): 875. DOI: 10.3390/cells9040875. |
| [9] |
KISSELEVA T, BRENNER D. Molecular and cellular mechanisms of liver fibrosis and its regression[J]. Nat Rev Gastroenterol Hepatol, 2021, 18(3): 151-166. DOI: 10.1038/s41575-020-00372-7. |
| [10] |
LEVINE B, KROEMER G. Biological functions of autophagy genes: a disease perspective[J]. Cell, 2019, 176(1-2): 11-42. DOI: 10.1016/j.cell.2018.09.048. |
| [11] |
WANG H, LIU Y, WANG D, et al. The upstream pathway of mTOR-mediated autophagy in liver diseases[J]. Cells, 2019, 8(12): 1597. DOI: 10.3390/cells8121597. |
| [12] |
LEE KW, THIYAGARAJAN V, SIE HW, et al. Synergistic effect of natural compounds on the fatty acid-induced autophagy of activated hepatic stellate cells[J]. J Nutr Biochem, 2014, 25(9): 903-913. DOI: 10.1016/j.jnutbio.2014.04.001. |
| [13] |
PARK M, KIM YH, WOO SY, et al. Tonsil-derived mesenchymal stem cells ameliorate CCl 4-induced liver fibrosis in mice via autophagy activation[J]. Sci Rep, 2015, 5: 8616. DOI: 10.1038/srep08616. |


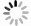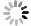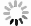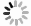





 DownLoad:
DownLoad: