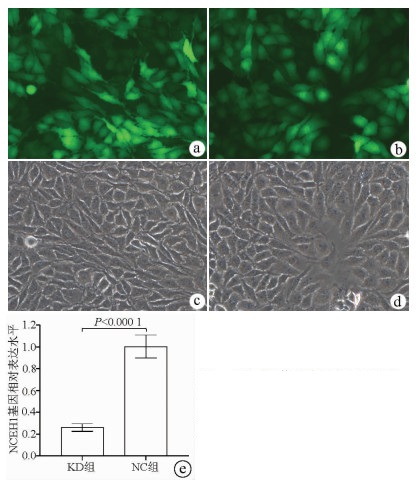
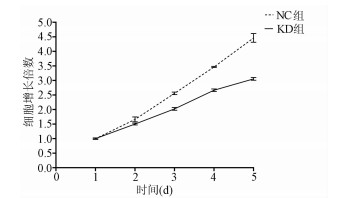
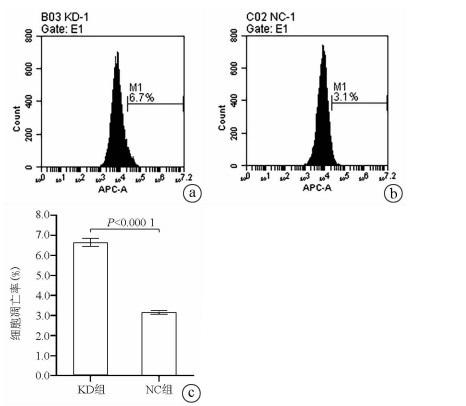
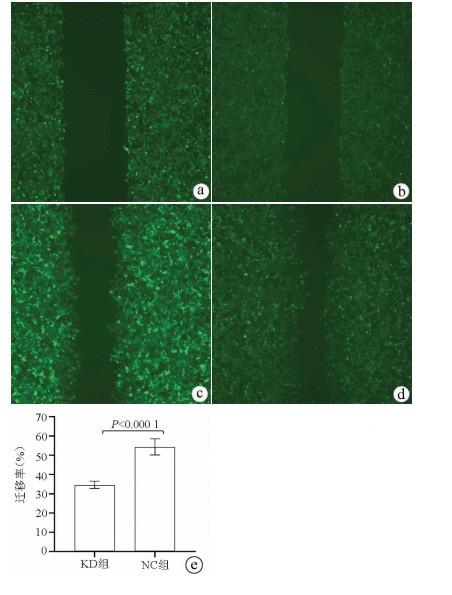
| [1] |
CHEN H, JIA W. Progress in hepatectomy for hepatocellular carcinoma and peri-operation management[J]. Genes Dis, 2020, 7(3): 320-327. DOI: 10.1016/j.gendis.2020.02.001. |
| [2] |
ALI ES, RYCHKOV GY, BARRITT GJ. Deranged hepatocyte intracellular Ca 2+ homeostasis and the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma[J]. Cell Calcium, 2019, 82: 102057. DOI: 10.1016/j.ceca.2019.102057. |
| [3] |
ALY SM, FETAIH HA, HASSANIN A, et al. Protective effects of garlic and cinnamon oils on hepatocellular carcinoma in albino rats[J]. Anal Cell Pathol (Amst), 2019, 2019: 9895485. DOI: 10.1155/2019/9895485. |
| [4] |
IGARASHI M, OSUGA J, UOZAKI H, et al. The critical role of neutral cholesterol ester hydrolase 1 in cholesterol removal from human macrophages[J]. Circ Res, 2010, 107(11): 1387-1395. DOI: 10.1161/CIRCRESAHA.110.226613. |
| [5] |
CHANG JW, NOMURA DK, CRAVATT BF. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis[J]. Chem Biol, 2011, 18(4): 476-484. DOI: 10.1016/j.chembiol.2011.02.008. |
| [6] |
SHI WJ, LI XC, WEN HX, et al. Bioinformatics analysis of AL360181.1 regulating the progression and prognosis of colorectal cancer[J]. J Xi'an Univ (Med Sci), 2020, 41(5): 731-737. DOI: 10.7652/jdyxb202005018. 史维俊, 李欣灿, 温贺新, 等. 基因AL360181.1调节结直肠癌进展及预后的生物信息学分析[J]. 西安交通大学学报(医学版), 2020, 41(5): 731-737. DOI: 10.7652/jdyxb202005018. |
| [7] |
YE Z, WANG S, CHEN W, et al. Fat mass and obesity-associated protein promotes the tumorigenesis and development of liver cancer[J]. Oncol Lett, 2020, 20(2): 1409-1417. DOI: 10.3892/ol.2020.11673. |
| [8] |
LI P, LIU Y. Effect of hepatitis B x gene on the expression of major histocompatibility complex class Ⅰ chain-related gene A-A5.1, invasion, and migration of HepG2.2.15 cells[J]. J Clin Hepatol, 2020, 36(4): 808-812. DOI: 10.3969/j.issn.1001-5256.2020.04.020. |
| [9] |
HO SY, HSU CY, LIU PH, et al. Albumin-bilirubin (ALBI) grade-based nomogram to predict tumor recurrence in patients with hepatocellular carcinoma[J]. Eur J Surg Oncol, 2019, 45(5): 776-781. DOI: 10.1016/j.ejso.2018.10.541. |
| [10] |
|
| [11] |
JIANG Y, SUN A, ZHAO Y, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma[J]. Nature, 2019, 567(7747): 257-261. DOI: 10.1038/s41586-019-0987-8. |
| [12] |
LIN L, DING Y, WANG Y, et al. Functional lipidomics: Palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism[J]. Hepatology, 2017, 66(2): 432-448. DOI: 10.1002/hep.29033. |
| [13] |
POPE ED 3rd, KIMBROUGH EO, VEMIREDDY LP, et al. Aberrant lipid metabolism as a therapeutic target in liver cancer[J]. Expert Opin Ther Targets, 2019, 23(6): 473-483. DOI: 10.1080/14728222.2019.1615883. |
| [14] |
NAKAGAWA H, HAYATA Y, KAWAMURA S, et al. Lipid metabolic reprogramming in hepatocellular carcinoma[J]. Cancers (Basel), 2018, 10(11): 447. DOI: 10.3390/cancers10110447. |
| [15] |
CHISTIAKOV DA, BOBRYSHEV YV, OREKHOV AN. Macrophage-mediated cholesterol handling in atherosclerosis[J]. J Cell Mol Med, 2016, 20(1): 17-28. DOI: 10.1111/jcmm.12689. |
| [16] |
LUCAS EK, DOUGHERTY SE, MCMEEKIN LJ, et al. PGC-1α provides a transcriptional framework for synchronous neurotransmitter release from parvalbumin-positive interneurons[J]. J Neurosci, 2014, 34(43): 14375-14387. DOI: 10.1523/JNEUROSCI.1222-14.2014. |
| [17] |
CHIANG KP, NIESSEN S, SAGHATELIAN A, et al. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling[J]. Chem Biol, 2006, 13(10): 1041-1050. DOI: 10.1016/j.chembiol.2006.08.008. |
| [18] |
NOMURA DK, FUJIOKA K, ISSA RS, et al. Dual roles of brain serine hydrolase KIAA1363 in ether lipid metabolism and organophosphate detoxification[J]. Toxicol Appl Pharmacol, 2008, 228(1): 42-48. DOI: 10.1016/j.taap.2007.11.021. |
| [19] |
CHEUNG L, FISHER RM, KUZMINA N, et al. Psoriasis skin inflammation-induced microRNA-26b targets NCEH1 in underlying subcutaneous adipose tissue[J]. J Invest Dermatol, 2016, 136(3): 640-648. DOI: 10.1016/j.jid.2015.12.008. |
| [20] |
ZHOU A, PAN H, SUN D, et al. miR-26b-5p inhibits the proliferation, migration and invasion of human papillary thyroid cancer in a β-catenin-dependent manner[J]. Onco Targets Ther, 2020, 13: 1593-1603. DOI: 10.2147/OTT.S236319. |
| [21] |
WANG Y, SUN B, ZHAO X, et al. Twist1-related miR-26b-5p suppresses epithelial-mesenchymal transition, migration and invasion by targeting SMAD1 in hepatocellular carcinoma[J]. Oncotarget, 2016, 7(17): 24383-24401. DOI: 10.18632/oncotarget.8328. |
| [22] |
KHOSLA R, HEMATI H, RASTOGI A, et al. miR-26b-5p helps in EpCAM+cancer stem cells maintenance via HSC71/HSPA8 and augments malignant features in HCC[J]. Liver Int, 2019, 39(9): 1692-1703. DOI: 10.1111/liv.14188. |
| [23] |
GUPTA M, CHANDAN K, SARWAT M. Role of microRNA and long non-coding RNA in hepatocellular carcinoma[J]. Curr Pharm Des, 2020, 26(4): 415-428. DOI: 10.2174/1381612826666200115093835. |






 DownLoad:
DownLoad: