| [1] |
CRAIG AJ, VON FELDEN J, GARCIA-LEZANA T, et al. Tumour evolution in hepatocellular carcinoma[J]. Nat Rev Gastroenterol Hepatol, 2020, 17(3): 139-152. DOI: 10.1038/s41575-019-0229-4. |
| [2] |
BRAY F, FERLAY J, SOERJOMATARAM I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68(6): 394-424. DOI: 10.3322/caac.21492. |
| [3] |
BRUIX J, REIG M, SHERMAN M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma[J]. Gastroenterology, 2016, 150(4): 835-853. DOI: 10.1053/j.gastro.2015.12.041. |
| [4] |
WU Y, LIU Z, XU X. Molecular subtyping of hepatocellular carcinoma: A step toward precision medicine[J]. Cancer Commun (Lond), 2020, 40(12): 681-693. DOI: 10.1002/cac2.12115. |
| [5] |
GONG J, CHEHRAZI-RAFFLE A, REDDI S, et al. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations[J]. J Immunother Cancer, 2018, 6(1): 8. DOI: 10.1186/s40425-018-0316-z. |
| [6] |
LLOVET JM, ZUCMAN-ROSSI J, PIKARSKY E, et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2016, 2: 16018. DOI: 10.1038/nrdp.2016.18. |
| [7] |
LLOVET JM, BRU' C, BRUIX J. Prognosis of hepatocellular carcinoma: The BCLC staging classification[J]. Semin Liver Dis, 1999, 19(3): 329-338. DOI: 10.1055/s-2007-1007122. |
| [8] |
BANNASCH P, RIBBACK S, SU Q, et al. Clear cell hepatocellular carcinoma: Origin, metabolic traits and fate of glycogenotic clear and ground glass cells[J]. Hepatobiliary Pancreat Dis Int, 2017, 16(6): 570-594. DOI: 10.1016/S1499-3872(17)60071-7. |
| [9] |
FAIVRE S, RIMASSA L, FINN RS. Molecular therapies for HCC: Looking outside the box[J]. J Hepatol, 2020, 72(2): 342-352. DOI: 10.1016/j.jhep.2019.09.010. |
| [10] |
ERSTAD DJ, FUCHS BC, TANABE KK. Molecular signatures in hepatocellular carcinoma: A step toward rationally designed cancer therapy[J]. Cancer, 2018, 124(15): 3084-3104. DOI: 10.1002/cncr.31257. |
| [11] |
CHIANG DY, VILLANUEVA A, HOSHIDA Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma[J]. Cancer Res, 2008, 68(16): 6779-6788. DOI: 10.1158/0008-5472.CAN-08-0742. |
| [12] |
SCHULZE K, IMBEAUD S, LETOUZÉ E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets[J]. Nat Genet, 2015, 47(5): 505-511. DOI: 10.1038/ng.3252. |
| [13] |
FUJIMOTO A, FURUTA M, TOTOKI Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer[J]. Nat Genet, 2016, 48(5): 500-509. DOI: 10.1038/ng.3547. |
| [14] |
LEE JS, CHU JS, CHU IS, HEO J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling[J]. Hepatology, 2004, 40(3): 667-676. DOI: 10.1002/hep.20375. |
| [15] |
JIANG X, KIM HE, SHU H, et al. Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway[J]. Science, 2003, 299(5604): 223-226. DOI: 10.1126/science.1076807. |
| [16] |
SEMENZA GL. Targeting HIF-1 for cancer therapy[J]. Nat Rev Cancer, 2003, 3(10): 721-732. DOI: 10.1038/nrc1187. |
| [17] |
LEE JS, HEO J, LIBBRECHT L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells[J]. Nat Med, 2006, 12(4): 410-416. DOI: 10.1038/nm1377. |
| [18] |
BOYAULT S, RICKMAN DS, de REYNIÈS A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets[J]. Hepatology, 2007, 45(1): 42-52. DOI: 10.1002/hep.21467. |
| [19] |
HOSHIDA Y, NIJMAN SM, KOBAYASHI M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma[J]. Cancer Res, 2009, 69(18): 7385-7392. DOI: 10.1158/0008-5472.CAN-09-1089. |
| [20] |
ZAVADIL J, BÖTTINGER EP. TGF-beta and epithelial-to-mesenchymal transitions[J]. Oncogene, 2005, 24(37): 5764-5774. DOI: 10.1038/sj.onc.1208927. |
| [21] |
GIANNELLI G, BERGAMINI C, FRANSVEA E, et al. Laminin-5 with transforming growth factor-beta1 induces epithelial to mesenchymal transition in hepatocellular carcinoma[J]. Gastroenterology, 2005, 129(5): 1375-1383. DOI: 10.1053/j.gastro.2005.09.055. |
| [22] |
HOPPLER S, KAVANAGH CL. Wnt signalling: Variety at the core[J]. J Cell Sci, 2007, 120(Pt 3): 385-393. DOI: 10.1242/jcs.03363. |
| [23] |
MUTO Y, MORIWAKI H, SHIRATORI Y. Prevention of second primary tumors by an acyclic retinoid, polyprenoic acid, in patients with hepatocellular carcinoma[J]. Digestion, 1998, 59(Suppl 2): 89-91. DOI: 10.1159/000051435. |
| [24] |
ZHANG Y, WANG G, MA W, et al. CdS p-n heterojunction co-boosting with Co(3)O(4) and Ni-MOF-74 for photocatalytic hydrogen evolution[J]. Dalton Trans, 2018, 47(32): 11176-11189. DOI: 10.1039/c8dt02294a. |
| [25] |
HANAHAN D, WEINBERG RA. Hallmarks of cancer: The next generation[J]. Cell, 2011, 144(5): 646-674. DOI: 10.1016/j.cell.2011.02.013. |
| [26] |
DÉSERT R, ROHART F, CANAL F, et al. Human hepatocellular carcinomas with a periportal phenotype have the lowest potential for early recurrence after curative resection[J]. Hepatology, 2017, 66(5): 1502-1518. DOI: 10.1002/hep.29254. |
| [27] |
YANG C, HUANG X, LIU Z, et al. Metabolism-associated molecular classification of hepatocellular carcinoma[J]. Mol Oncol, 2020, 14(4): 896-913. DOI: 10.1002/1878-0261.12639. |
| [28] |
SHANKARAIAH RC, CALLEGARI E, GUERRIERO P, et al. Metformin prevents liver tumourigenesis by attenuating fibrosis in a transgenic mouse model of hepatocellular carcinoma[J]. Oncogene, 2019, 38(45): 7035-7045. DOI: 10.1038/s41388-019-0942-z. |
| [29] |
DUFFY AG, ULAHANNAN SV, MAKOROVA-RUSHER O, et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma[J]. J Hepatol, 2017, 66(3): 545-551. DOI: 10.1016/j.jhep.2016.10.029. |
| [30] |
EL-KHOUEIRY AB, SANGRO B, YAU T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial[J]. Lancet, 2017, 389(10088): 2492-2502. DOI: 10.1016/S0140-6736(17)31046-2. |
| [31] |
KILLOCK D. Immunotherapy: Nivolumab keeps HCC in check and opens avenues for checkmate[J]. Nat Rev Clin Oncol, 2017, 14(7): 392. DOI: 10.1038/nrclinonc.2017.70. |
| [32] |
JIANG Y, SUN A, ZHAO Y, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma[J]. Nature, 2019, 567(7747): 257-261. DOI: 10.1038/s41586-019-0987-8. |
| [33] |
GAO Q, ZHU H, DONG L, et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma[J]. Cell, 2019, 179(2): 561-577. e22. DOI: 10.1016/j.cell.2019.08.052. |
| [34] |
SIA D, JIAO Y, MARTINEZ-QUETGLAS I, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features[J]. Gastroenterology, 2017, 153(3): 812-826. DOI: 10.1053/j.gastro.2017.06.007. |
| [35] |
KUREBAYASHI Y, OJIMA H, TSUJIKAWA H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification[J]. Hepatology, 2018, 68(3): 1025-1041. DOI: 10.1002/hep.29904. |
| [36] |
KIM HD, SONG GW, PARK S, et al. Association between expression level of PD1 by tumor-infiltrating CD8 +T cells and features of hepatocellular carcinoma[J]. Gastroenterology, 2018, 155(6): 1936-1950. e17. DOI: 10.1053/j.gastro.2018.08.030. |
| [37] |
ZHANG Q, LOU Y, YANG J, et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas[J]. Gut, 2019, 68(11): 2019-2031. DOI: 10.1136/gutjnl-2019-318912. |
| [38] |
LIM HY, HEO J, CHOI HJ, et al. A phase Ⅱ study of the efficacy and safety of the combination therapy of the MEK inhibitor refametinib (BAY 86-9766) plus sorafenib for Asian patients with unresectable hepatocellular carcinoma[J]. Clin Cancer Res, 2014, 20(23): 5976-5985. DOI: 10.1158/1078-0432.CCR-13-3445. |
| [39] |
HO DWH, CHAN LK, CHIU YT, et al. TSC1/2 mutations define a molecular subset of HCC with aggressive behaviour and treatment implication[J]. Gut, 2017, 66(8): 1496-1506. DOI: 10.1136/gutjnl-2016-312734. |
| [40] |
SUN W, LI SC, XU L, et al. High FLT3 levels may predict sorafenib benefit in hepatocellular carcinoma[J]. Clin Cancer Res, 2020, 26(16): 4302-4312. DOI: 10.1158/1078-0432.CCR-19-1858. |


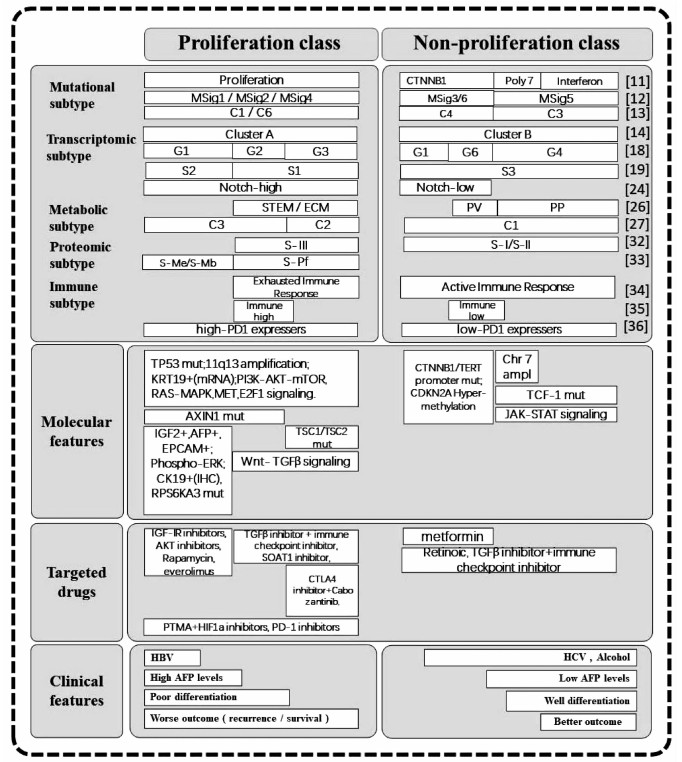




 DownLoad:
DownLoad: