| [1] |
DUTTA R, MAHATO RI. Recent advances in hepatocellular carcinoma therapy[J]. Pharmacol Ther, 2017, 173(5): 106-117.
|
| [2] |
ZHANG Z, MA L, GOSWAMI S, et al. Landscape of infiltrating B cells and their clinical significance in human hepatocellular carcinoma[J]. OncoImmunology, 2019, 8(4): 1-11.
|
| [3] |
ZHAO ZJ, HU Y, SHEN XH, et al. HBx represses RIZ1 expression by DNA methyltransferase 1 involvement in decreased miR-152 in hepatocellular carcinoma[J]. Oncol Rep, 2017, 37(5): 2811. DOI: 10.3892/or.2017.5518 |
| [4] |
DUAN YF, ZHAO QX, JING X. Effect of long non-coding RNAs on glycolytic pathway in primary liver cancer and related mechanisms[J]. J Clin Hepatol, 2019, 35(6): 1374-1376. (in Chinese) DOI: 10.3969/j.issn.1001-5256.2019.06.043 |
| [5] |
|
| [6] |
|
| [7] |
ZHOU JJ, ZHANG YJ, QI YH, et al. MicroRNA-152 inhibits tumor cell growth by directly targeting RTKN in hepatocellular carcinoma[J]. Oncol Rep, 2017, 37(2): 1227. DOI: 10.3892/or.2016.5290 |
| [8] |
MIQUELESTORENA-STANDLEY E, TALLET A, COLLIN C, et al. Interest of variations in microRNA-152 and-122 in a series of hepatocellular carcinomas related to hepatitis C virus infection[J]. Hepatol Res, 2018, 48(7): 566-573. DOI: 10.1111/hepr.12915 |
| [9] |
BILIC′ P, GUILLEMIN N, KOVACˇEVIC′ A, et al. Serum proteome profiling in canine idiopathic dilated cardiomyopathy using TMT-based quantitative proteomics approach[J]. J Proteomics, 2018, 179: 110-121. DOI: 10.1016/j.jprot.2018.03.007 |
| [10] |
|
| [11] |
CHEN L, ZHANG YH, WANG SP, et al. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways[J]. PLoS One, 2017, 12(9): e0184129. DOI: 10.1371/journal.pone.0184129 |
| [12] |
WANG F, YING H, HE B, et al. Circulating miR-148/152 family as potential biomarkers in hepatocellular carcinoma[J]. Tumour Biol, 2016, 37(4): 4945-4953. DOI: 10.1007/s13277-015-4340-z |
| [13] |
HUANG J, WANG Y, GUO Y, et al. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1[J]. Hepatology, 2010, 52(1): 60-70. DOI: 10.1002/hep.23660 |
| [14] |
DANG YW, ZENG J, HE RQ, et al. Effects of miR-152 on cell growth inhibition, motility suppression and apoptosis induction in hepatocellular carcinoma cells[J]. Asian Pac J Cancer Prev, 2014, 15(12): 4969-4976. DOI: 10.7314/APJCP.2014.15.12.4969 |
| [15] |
IRYNA K, VOLODYMYR T, ALINE DC, et al. MicroRNA-152-mediated dysregulation of hepatic transferrin receptor 1 in liver carcinogenesis[J]. Oncotarget, 2016, 7(2): 1276-1287. DOI: 10.18632/oncotarget.6004 |
| [16] |
FORMOSA LE, MASAKAZU M, FRAZIER AE, et al. Characterization of mitochondrial FOXRED1 in the assembly of respiratory chain complex I[J]. Hum Mol Genet, 2015, 24(10): 2952-2965. DOI: 10.1093/hmg/ddv058 |
| [17] |
DAGDA RK, GUSDON AM, PIEN I, et al. Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson's disease[J]. Cell Death Differ, 2011, 18(12): 1914-1923. DOI: 10.1038/cdd.2011.74 |
| [18] |
YU J, ZHANG Y, ZHOU DX, et al. Higher expression of A-kinase anchoring-protein 1 predicts poor prognosis in human hepatocellular carcinoma[J]. Oncol Lett, 2018, 16(1): 131-136.
|
| [19] |
|
| [20] |
ZHANG XC, LI B. Towards understanding the mechanisms of proton pumps in Complex-Ⅰ of the respiratory chain[J]. Biophy Rep, 2019, 5(5-6): 219-234. DOI: 10.1007/s41048-019-00094-7 |


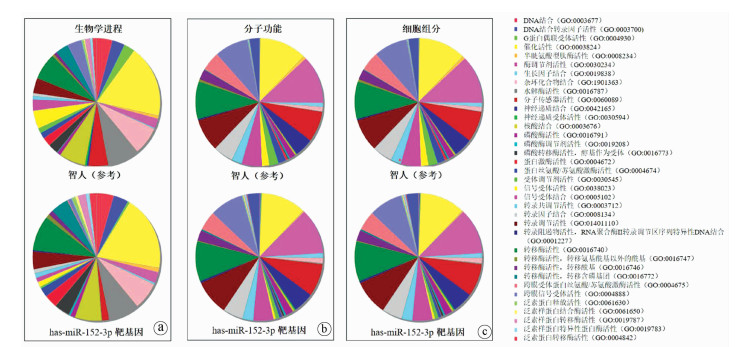




 DownLoad:
DownLoad: