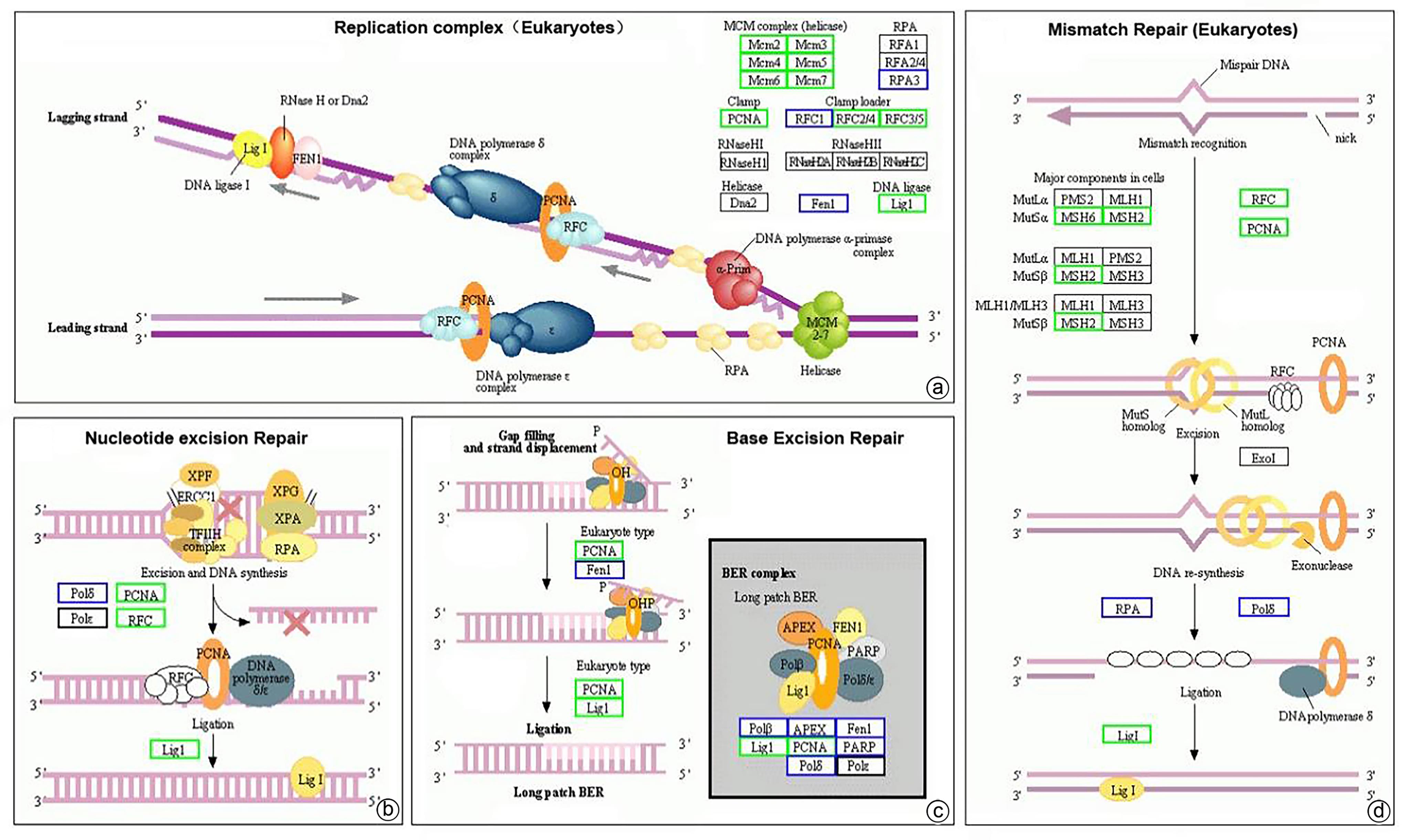
| [1] |
HU ZQ, YIN YF, JIANG J, et al. Exosomal miR-142-3p secreted by hepatitis B virus(HBV)-hepatocellular carcinoma(HCC) cells promotes ferroptosis of M1-type macrophages through SLC3A2 and the mechanism of HCC progression[J]. J Gastrointest Oncol, 2022, 13( 2): 754- 767. DOI: 10.21037/jgo-21-916. |
| [2] |
KATO K, FUKAI M, HATANAKA KC, et al. Versican secreted by cancer-associated fibroblasts is a poor prognostic factor in hepatocellular carcinoma[J]. Ann Surg Oncol, 2022, 29( 11): 7135- 7146. DOI: 10.1245/s10434-022-11862-0. |
| [3] |
PANG SJ, SHI Y, XU DP, et al. Screening of hepatocellular carcinoma patients with high risk of early recurrence after radical hepatectomy using a nomogram model based on the γ-glutamyl transpeptidase-to-albumin ratio[J]. J Gastrointest Surg, 2022, 26( 8): 1- 9. DOI: 10.1007/s11605-022-05326-9. |
| [4] |
GOMEZ-QUIROZ LE, ROMAN S. Influence of genetic and environmental risk factors in the development of hepatocellular carcinoma in Mexico[J]. Ann Hepatol, 2022, 27( Suppl 1): 100649. DOI: 10.1016/j.aohep.2021.100649. |
| [5] |
LUO MJ, ZHAO YX, WANG YD, et al. Comparative proteomics of contrasting maize genotypes provides insights into salt-stress tolerance mechanisms[J]. J Proteome Res, 2018, 17( 1): 141- 153. DOI: 10.1021/acs.jproteome.7b00455. |
| [6] |
SHRESTHA P, KIM MS, ELBASANI E, et al. Prediction of trehalose-metabolic pathway and comparative analysis of KEGG, MetaCyc, and RAST databases based on complete genome of Variovorax sp. PAMC28711[J]. BMC Genom Data, 2022, 23( 1): 4. DOI: 10.1186/s12863-021-01020-y. |
| [7] |
MURANUSHI R, ARAKI K, YOKOBORI T, et al. High membrane expression of CMTM6 in hepatocellular carcinoma is associated with tumor recurrence[J]. Cancer Sci, 2021, 112( 8): 3314- 3323. DOI: 10.1111/cas.15004. |
| [8] |
ZHONG F, XIE C, PENG X, et al. A commentary on‘Laparoscopic versus open repeat hepatectomy for recurrent hepatocellular carcinoma: A systematic review and meta-analysis of propensity score-matched cohort studies’[J]. Int J Surg, 2023, 109( 9): 2821- 2822. DOI: 10.1097/js9.0000000000000517. |
| [9] |
BAI YP, SHA JJ, OKUI T, et al. The epithelial-mesenchymal transition influences the resistance of oral squamous cell carcinoma to monoclonal antibodies via its effect on energy homeostasis and the tumor microenvironment[J]. Cancers, 2021, 13( 23): 5905. DOI: 10.3390/cancers13235905. |
| [10] |
HUANG DP, LIAO MM, TONG JJ, et al. Construction of a genome instability-derived lncRNA-based risk scoring system for the prognosis of hepatocellular carcinoma[J]. Aging, 2021, 13( 22): 24621- 24639. DOI: 10.18632/aging.203698. |
| [11] |
CHEN Y, HUANG MJ, ZHU JK, et al. Identification of a DNA damage response and repair-related gene-pair signature for prognosis stratification analysis in hepatocellular carcinoma[J]. Front Pharmacol, 2022, 13: 857060. DOI: 10.3389/fphar.2022.857060. |
| [12] |
BARCENA-VARELA M, LUJAMBIO A. A novel long noncoding RNA finetunes the DNA damage response in hepatocellular carcinoma[J]. Cancer Res, 2021, 81( 19): 4899- 4900. DOI: 10.1158/0008-5472.CAN-21-2776. |
| [13] |
WANG ZY, WANG XX, RONG ZH, et al. LncRNA LINC01134 contributes to radioresistance in hepatocellular carcinoma by regulating DNA damage response via MAPK signaling pathway[J]. Front Pharmacol, 2022, 12: 791889. DOI: 10.3389/fphar.2021.791889. |
| [14] |
LIU HH, YAN YC, CHEN RB, et al. Integrated nomogram based on five stage-related genes and TNM stage to predict 1-year recurrence in hepatocellular carcinoma[J]. Cancer Cell Int, 2020, 20: 140. DOI: 10.1186/s12935-020-01216-9. |
| [15] |
MARSHALL AE, RUSHBROOK SM, VOWLER SL, et al. Tumor recurrence following liver transplantation for hepatocellular carcinoma: Role of tumor proliferation status[J]. Liver Transpl, 2010, 16( 3): 279- 288. DOI: 10.1002/lt.21993. |
| [16] |
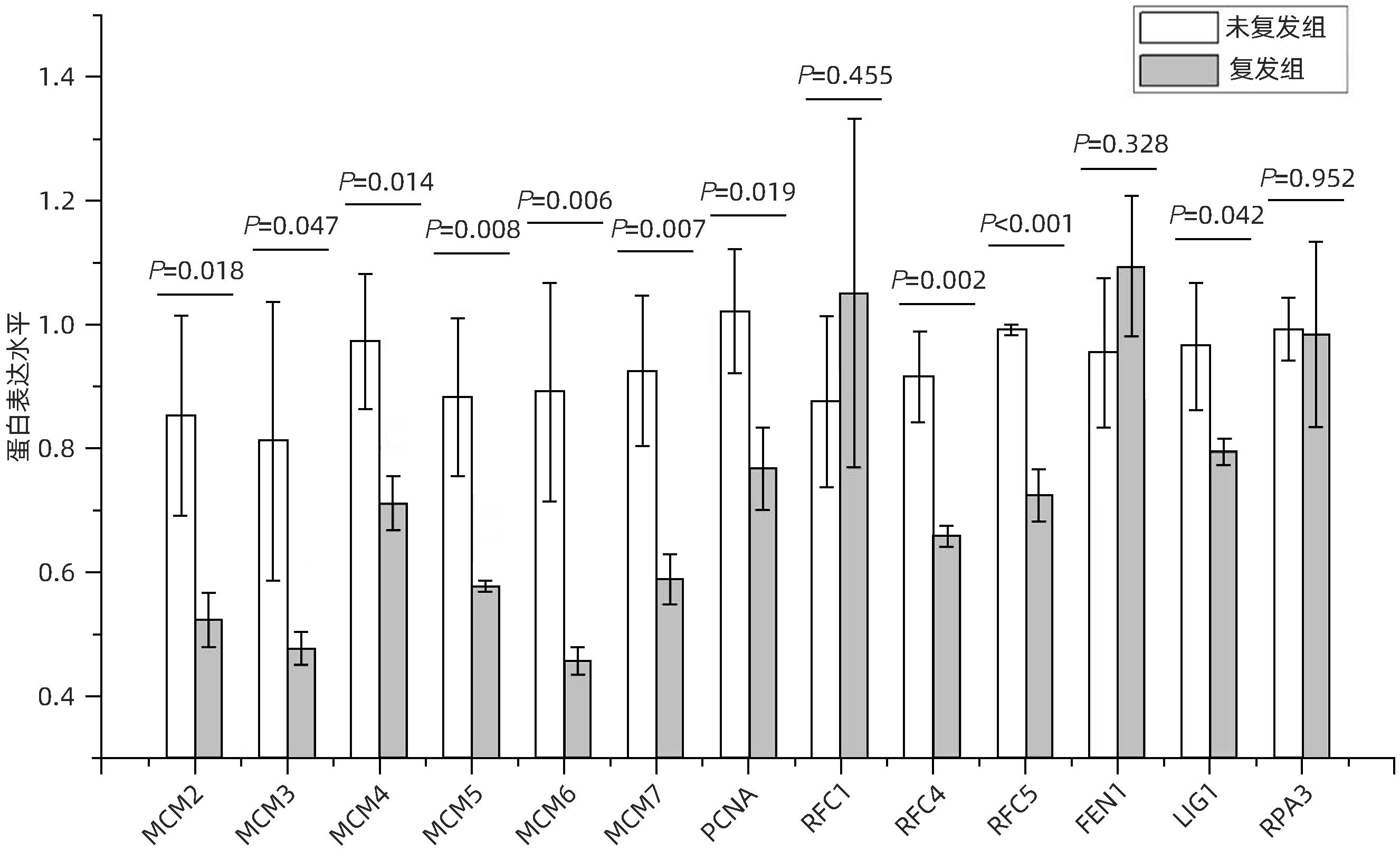
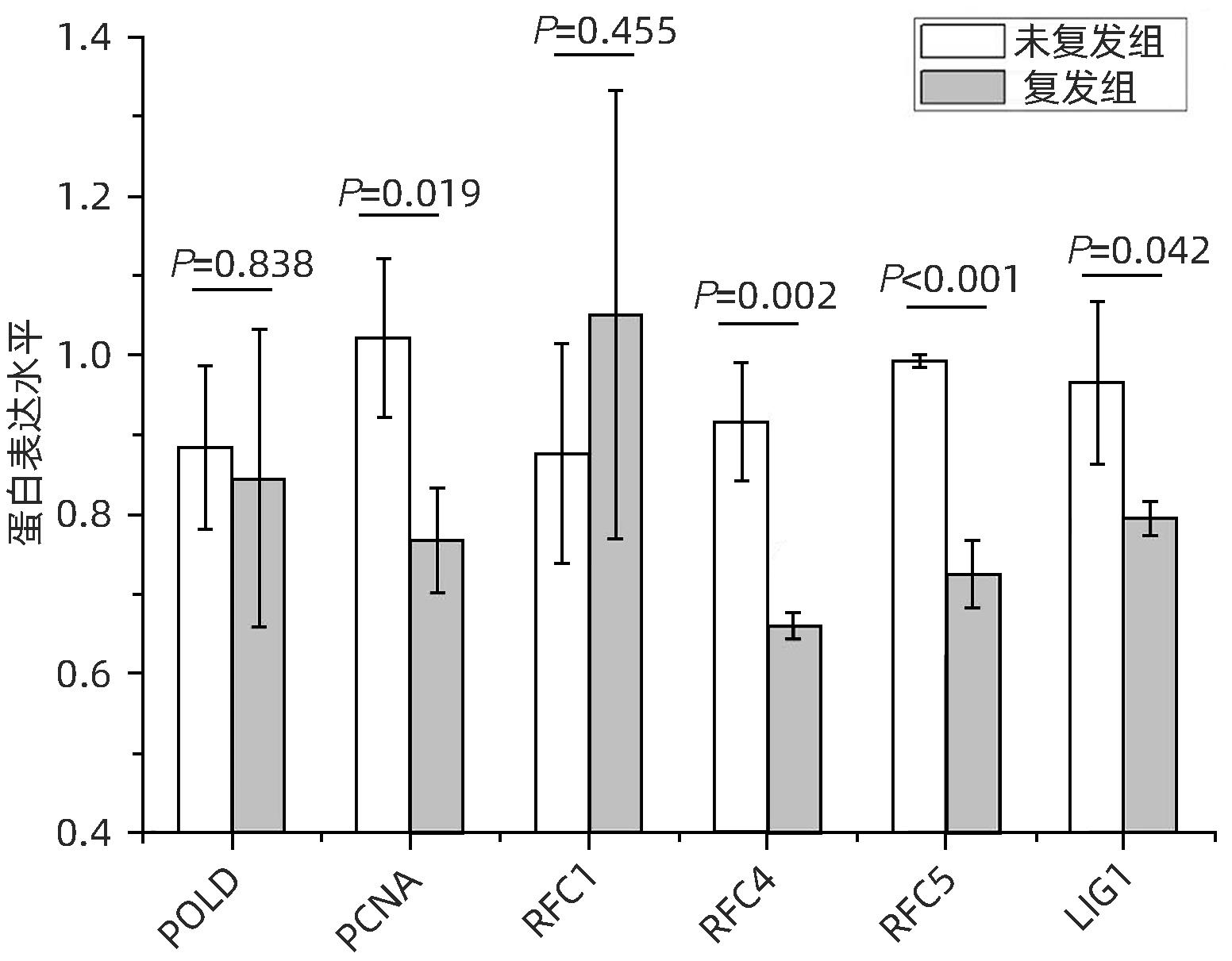
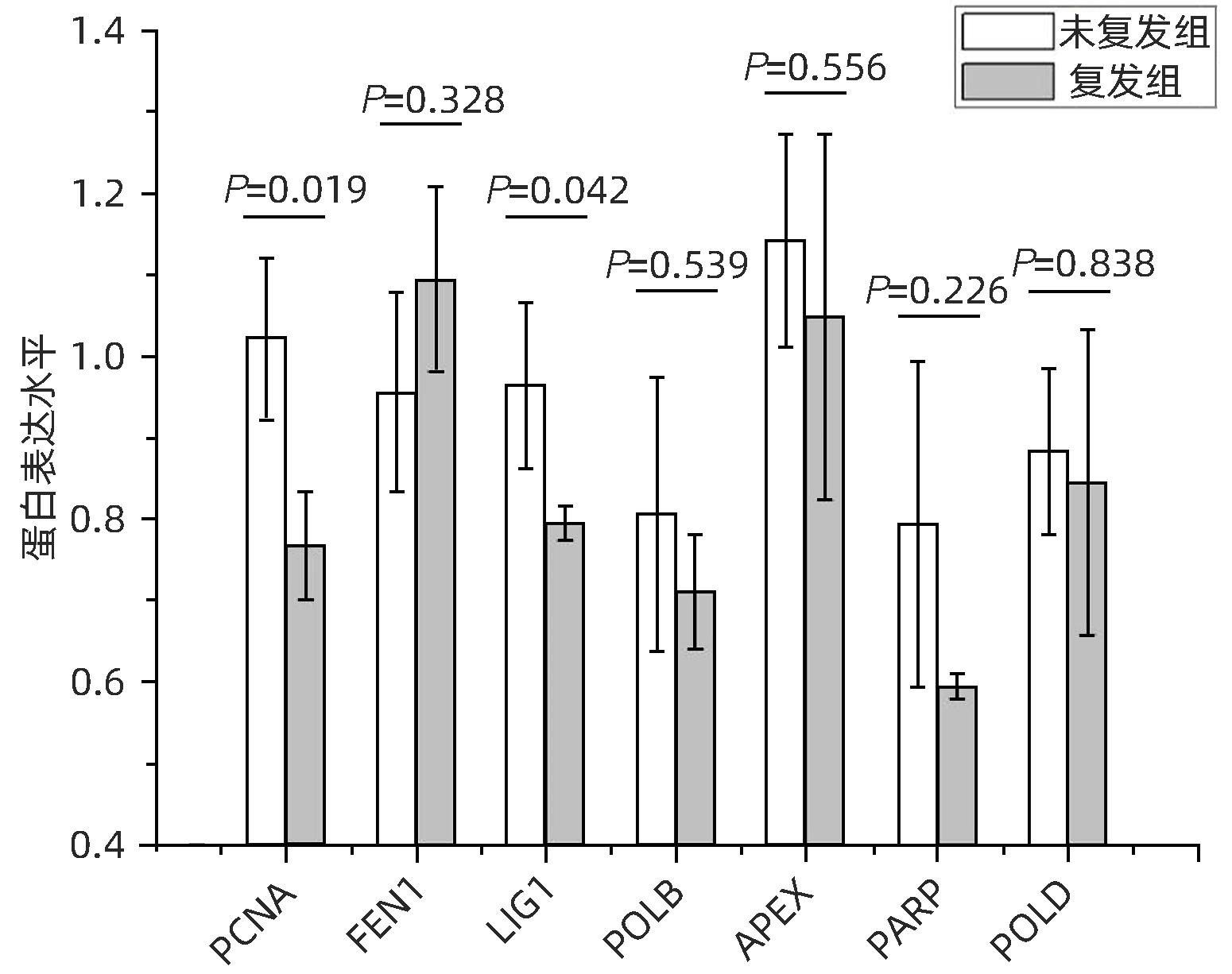
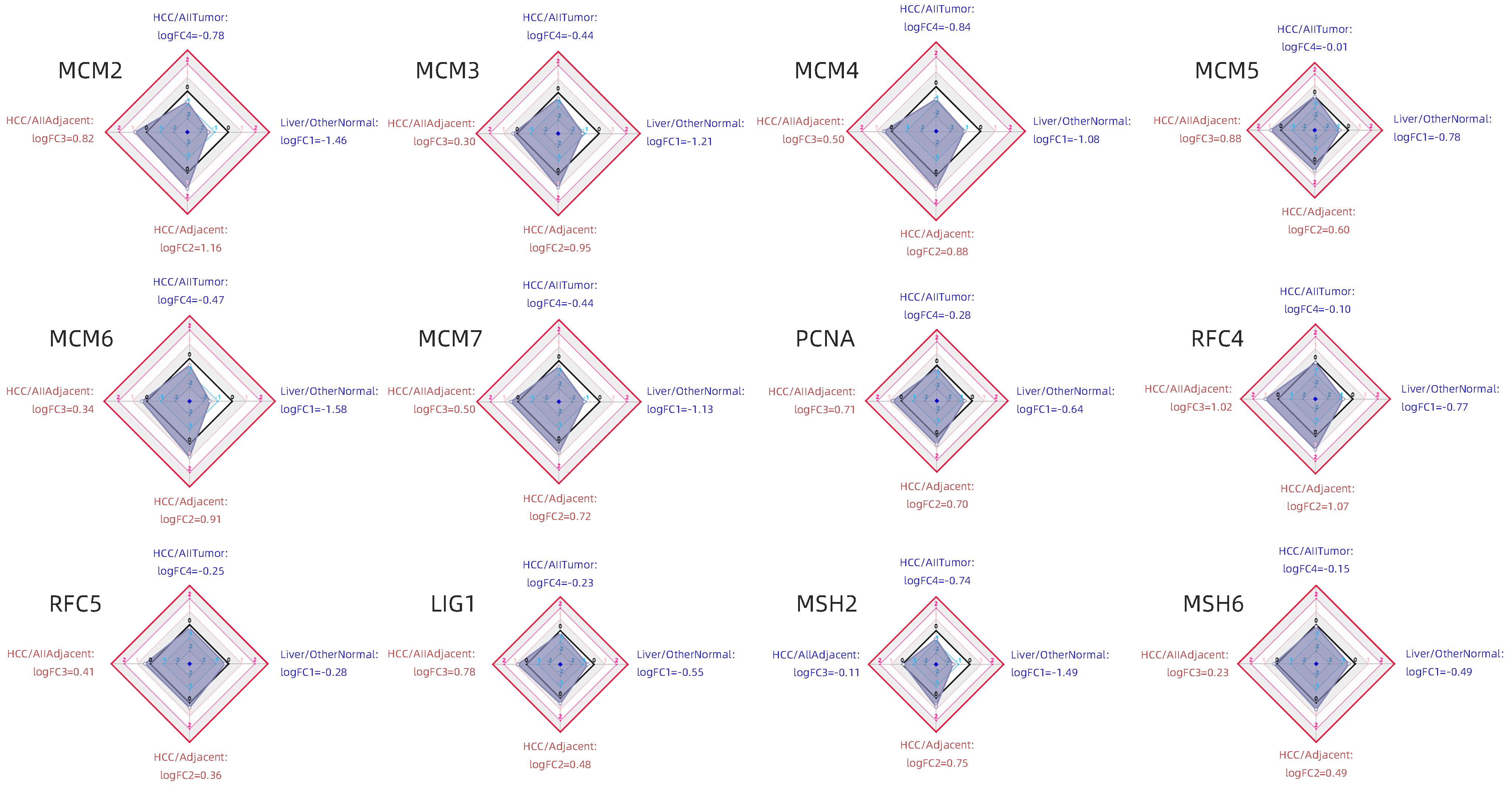
LIAO XW, LIU XG, YANG CK, et al. Distinct diagnostic and prognostic values of minichromosome maintenance gene expression in patients with hepatocellular carcinoma[J]. J Cancer, 2018, 9( 13): 2357- 2373. DOI: 10.7150/jca.25221. |
| [17] |
THUL PJ, LINDSKOG C. The human protein atlas: A spatial map of the human proteome[J]. Protein Sci, 2018, 27( 1): 233- 244. DOI: 10.1002/pro.3307. |


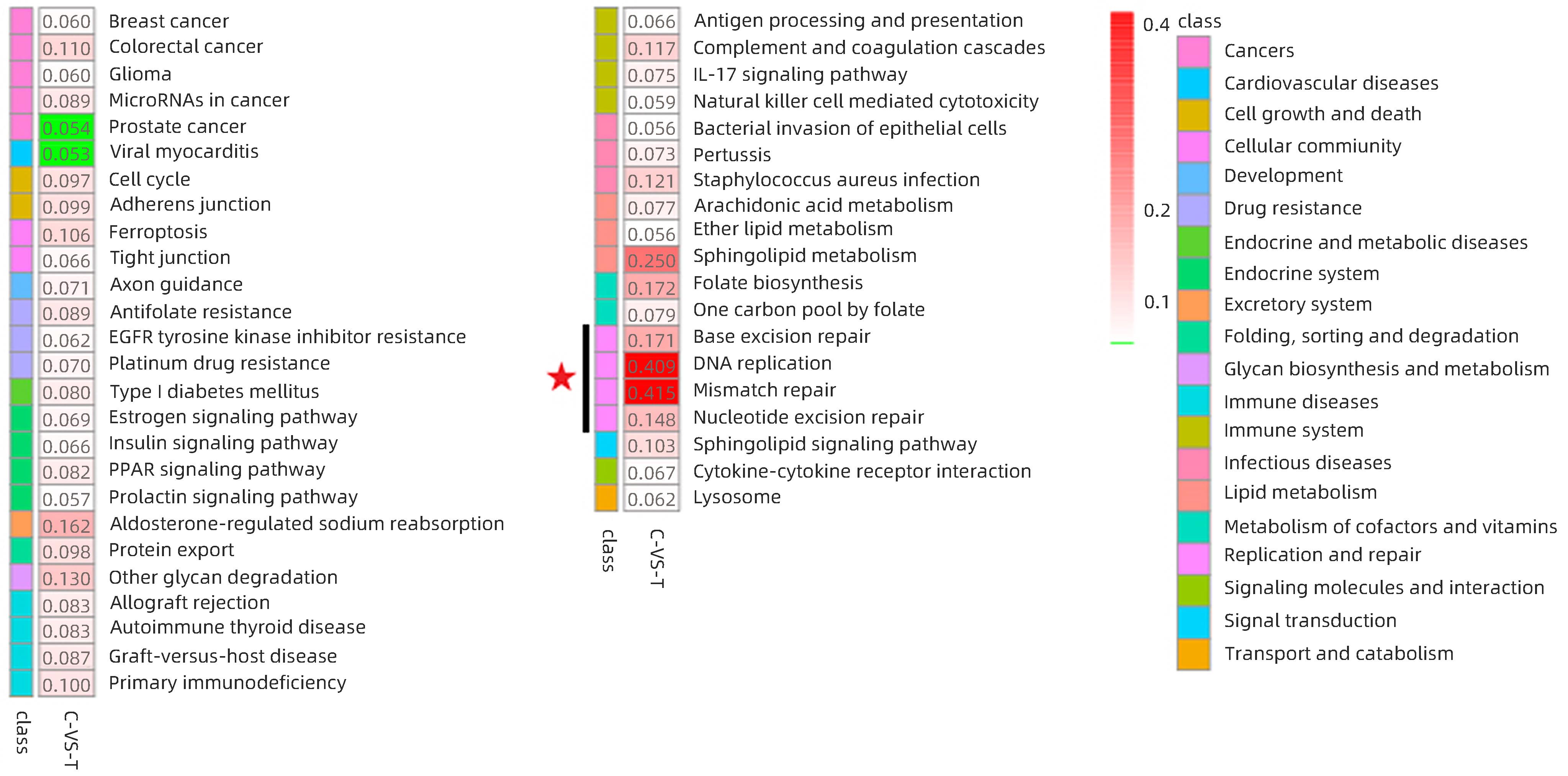




 DownLoad:
DownLoad: